The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) Genome: a high frequency of deletions in tandem duplicates
- PMID: 17507681
- PMCID: PMC1931530
- DOI: 10.1534/genetics.107.070805
The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) Genome: a high frequency of deletions in tandem duplicates
Abstract
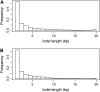
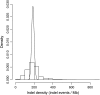
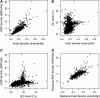
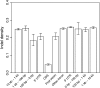
It is increasingly recognized that insertions and deletions (indels) are an important source of genetic as well as phenotypic divergence and diversity. We analyzed length polymorphisms identified through partial (0.25x) shotgun sequencing of three breeds of domestic chicken made by the International Chicken Polymorphism Map Consortium. A data set of 140,484 short indel polymorphisms in unique DNA was identified after filtering for microsatellite structures. There was a significant excess of tandem duplicates at indel sites, with deletions of a duplicate motif outnumbering the generation of duplicates through insertion. Indel density was lower in microchromosomes than in macrochromosomes, in the Z chromosome than in autosomes, and in 100 bp of upstream sequence, 5'-UTR, and first introns than in intergenic DNA and in other introns. Indel density was highly correlated with single nucleotide polymorphism (SNP) density. The mean density of indels in pairwise sequence comparisons was 1.9 x 10(-4) indel events/bp, approximately 5% the density of SNPs segregating in the chicken genome. The great majority of indels involved a limited number of nucleotides (median 1 bp), with A-rich motifs being overrepresented at indel sites. The overrepresentation of deletions at tandem duplicates indicates that replication slippage in duplicate sequences is a common mechanism behind indel mutation. The correlation between indel and SNP density indicates common effects of mutation and/or selection on the occurrence of indels and point mutations.
Figures
Similar articles
-
Indel segregating within introns in the chicken genome are positively correlated with the recombination rates.Hereditas. 2010 Apr;147(2):53-7. doi: 10.1111/j.1601-5223.2009.02141.x. Hereditas. 2010. PMID: 20536542
-
Genome-wide characterization of insertion and deletion variation in chicken using next generation sequencing.PLoS One. 2014 Aug 18;9(8):e104652. doi: 10.1371/journal.pone.0104652. eCollection 2014. PLoS One. 2014. PMID: 25133774 Free PMC article.
-
Detection and characterization of small insertion and deletion genetic variants in modern layer chicken genomes.BMC Genomics. 2015 Jul 31;16:562. doi: 10.1186/s12864-015-1711-1. BMC Genomics. 2015. PMID: 26227840 Free PMC article.
-
Research Progress on InDel Genetic Marker in Forensic Science.Fa Yi Xue Za Zhi. 2018 Aug;34(4):420-427. doi: 10.12116/j.issn.1004-5619.2018.04.016. Epub 2018 Aug 25. Fa Yi Xue Za Zhi. 2018. PMID: 30465411 Review. Chinese, English.
-
[DNA polymorphisms].Rinsho Byori. 2013 Nov;61(11):1001-7. Rinsho Byori. 2013. PMID: 24450105 Review. Japanese.
Cited by
-
Insertion-deletion polymorphisms (indels) as genetic markers in natural populations.BMC Genet. 2008 Jan 22;9:8. doi: 10.1186/1471-2156-9-8. BMC Genet. 2008. PMID: 18211670 Free PMC article.
-
Genome-wide resequencing of KRICE_CORE reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era.BMC Genomics. 2016 May 26;17:408. doi: 10.1186/s12864-016-2734-y. BMC Genomics. 2016. PMID: 27229151 Free PMC article.
-
The Impact of Natural Selection on Short Insertion and Deletion Variation in the Great Tit Genome.Genome Biol Evol. 2019 Jun 1;11(6):1514-1524. doi: 10.1093/gbe/evz068. Genome Biol Evol. 2019. PMID: 30924871 Free PMC article.
-
Evolutionary genomics of Colias Phosphoglucose Isomerase (PGI) introns.J Mol Evol. 2012 Feb;74(1-2):96-111. doi: 10.1007/s00239-012-9492-5. Epub 2012 Mar 3. J Mol Evol. 2012. PMID: 22388354
-
Artemisinin resistance in rodent malaria--mutation in the AP2 adaptor μ-chain suggests involvement of endocytosis and membrane protein trafficking.Malar J. 2013 Apr 5;12:118. doi: 10.1186/1475-2875-12-118. Malar J. 2013. PMID: 23561245 Free PMC article.
References
-
- Bhangale, T. R., M. J. Rieder, R. J. Livingston and D. A. Nickerson, 2005. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 14: 59–69. - PubMed
-
- Bois, P. R., 2003. Hypermutable minisatellites, a human affair? Genomics 81: 349–355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

