Contrasting effects of in vitro fertilization and nuclear transfer on the expression of mtDNA replication factors
- PMID: 17507682
- PMCID: PMC1931560
- DOI: 10.1534/genetics.106.070177
Contrasting effects of in vitro fertilization and nuclear transfer on the expression of mtDNA replication factors
Abstract
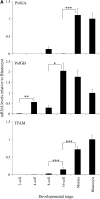
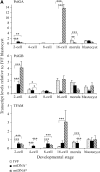
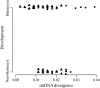
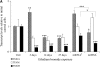
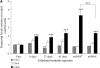
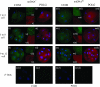
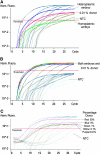
Mitochondrial DNA (mtDNA) is normally only inherited through the oocyte. However, nuclear transfer (NT), the fusion of a donor cell with an enucleated oocyte, can transmit both donor cell and recipient oocyte mtDNA. mtDNA replication is under the control of nuclear-encoded replication factors, such as polymerase gamma (POLG) and mitochondrial transcription factor A (TFAM). These are first expressed during late preimplantation embryo development. To account for the persistence of donor cell mtDNA, even when introduced at residual levels (mtDNA(R)), we hypothesized that POLG and TFAM would be upregulated in intra- and interspecific (ovine-ovine) and intergeneric (caprine-ovine) NT embryos when compared to in vitro fertilized (IVF) embryos. For the intra- and interspecific crosses, PolGA (catalytic subunit), PolGB (accessory subunit), and TFAM mRNA were expressed at the 2-cell stage in both nondepleted (mtDNA(+)) and mtDNA(R) embryos with protein being expressed up to the 16-cell stage for POLGA and TFAM. However, at the 16-cell stage, there was significantly more PolGA expression in the mtDNA(R) embryos compared to their mtDNA(+) counterparts. Expression for all three genes first matched IVF embryos at the blastocyst stage. In the intergeneric model, POLG was upregulated during preimplantation development. Although these embryos did not persist further than the 16+-cell stage, significantly more mtDNA(R) embryos reached this stage. However, the vast majority of these embryos were homoplasmic for recipient oocyte mtDNA. The upreglation in mtDNA replication factors was most likely due to the donor cells still expressing these factors prior to NT.
Figures





Similar articles
-
Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development.Biol Reprod. 2007 Feb;76(2):327-35. doi: 10.1095/biolreprod.106.054536. Epub 2006 Oct 11. Biol Reprod. 2007. PMID: 17035641
-
The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm.Hum Reprod. 2007 Jun;22(6):1585-96. doi: 10.1093/humrep/dem030. Epub 2007 Mar 5. Hum Reprod. 2007. PMID: 17339235
-
Expression of mitochondrial transcription factor A (TFAM) during porcine gametogenesis and preimplantation embryo development.J Cell Physiol. 2008 Nov;217(2):529-43. doi: 10.1002/jcp.21528. J Cell Physiol. 2008. PMID: 18636550
-
Nuclear transfer: preservation of a nuclear genome at the expense of its associated mtDNA genome(s).Curr Top Dev Biol. 2007;77:251-90. doi: 10.1016/S0070-2153(06)77010-7. Curr Top Dev Biol. 2007. PMID: 17222707 Review.
-
Role of mitochondrial DNA replication during differentiation of reprogrammed stem cells.Int J Dev Biol. 2010;54(11-12):1659-70. doi: 10.1387/ijdb.103202rk. Int J Dev Biol. 2010. PMID: 21404186 Review.
Cited by
-
Mitochondrial function of human embryo: Decline in their quality with maternal aging.Reprod Med Biol. 2022 Dec 21;21(1):e12491. doi: 10.1002/rmb2.12491. eCollection 2022 Jan-Dec. Reprod Med Biol. 2022. PMID: 36570768 Free PMC article. Review.
-
Irregular transcriptome reprogramming probably causes thec developmental failure of embryos produced by interspecies somatic cell nuclear transfer between the Przewalski's gazelle and the bovine.BMC Genomics. 2014 Dec 16;15(1):1113. doi: 10.1186/1471-2164-15-1113. BMC Genomics. 2014. PMID: 25511933 Free PMC article.
-
Interspecies nuclear transfer using fibroblasts from leopard, tiger, and lion ear piece collected postmortem as donor cells and rabbit oocytes as recipients.In Vitro Cell Dev Biol Anim. 2016 Jun;52(6):632-45. doi: 10.1007/s11626-016-0014-4. Epub 2016 Apr 12. In Vitro Cell Dev Biol Anim. 2016. PMID: 27071624
-
The effects of nuclear reprogramming on mitochondrial DNA replication.Stem Cell Rev Rep. 2013 Feb;9(1):1-15. doi: 10.1007/s12015-011-9318-7. Stem Cell Rev Rep. 2013. PMID: 21994000
-
Lack of effects of ooplasm transfer on early development of interspecies somatic cell nuclear transfer bison embryos.BMC Dev Biol. 2016 Oct 13;16(1):36. doi: 10.1186/s12861-016-0137-6. BMC Dev Biol. 2016. PMID: 27737629 Free PMC article.
References
-
- Anderson, S., A. T. Bankier, B. G. Barrell, M. H. De Bruijn, A. R. Coulson et al., 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. - PubMed
-
- Baguisi, A., E. Behboodi, D. T. Melican, J. S. Pollock, M. M. Destrempes et al., 1999. Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 17: 456–461. - PubMed
-
- Barrientos, A., L. Kenyon and C. T. Moraes, 1998. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 273: 14210–14217. - PubMed
-
- Bolton, V. N., P. J. Oades and M. H. Johnson, 1984. The relationship between cleavage, DNA replication, and gene expression in the mouse 2-cell embryo. J. Embryol. Exp. Morphol. 79: 139–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

