Multiple functions of Drosophila BLM helicase in maintenance of genome stability
- PMID: 17507683
- PMCID: PMC1950607
- DOI: 10.1534/genetics.106.070052
Multiple functions of Drosophila BLM helicase in maintenance of genome stability
Abstract
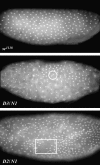
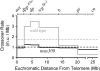
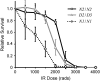
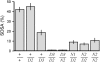
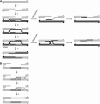
Bloom Syndrome, a rare human disorder characterized by genomic instability and predisposition to cancer, is caused by mutation of BLM, which encodes a RecQ-family DNA helicase. The Drosophila melanogaster ortholog of BLM, DmBlm, is encoded by mus309. Mutations in mus309 cause hypersensitivity to DNA-damaging agents, female sterility, and defects in repairing double-strand breaks (DSBs). To better understand these phenotypes, we isolated novel mus309 alleles. Mutations that delete the N terminus of DmBlm, but not the helicase domain, have DSB repair defects as severe as those caused by null mutations. We found that female sterility is due to a requirement for DmBlm in early embryonic cell cycles; embryos lacking maternally derived DmBlm have anaphase bridges and other mitotic defects. These defects were less severe for the N-terminal deletion alleles, so we used one of these mutations to assay meiotic recombination. Crossovers were decreased to about half the normal rate, and the remaining crossovers were evenly distributed along the chromosome. We also found that spontaneous mitotic crossovers are increased by several orders of magnitude in mus309 mutants. These results demonstrate that DmBlm functions in multiple cellular contexts to promote genome stability.
Figures

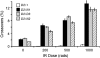
References
-
- Adams, M. D., and J. J. Sekelsky, 2002. From sequence to genotype: reverse genetics in Drosophila. Nat. Rev. Genet. 3: 189–198. - PubMed
-
- Adams, M. D., M. McVey and J. J. Sekelsky, 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. - PubMed
-
- Baker, B. S., and J. C. Hall, 1976. Meiotic mutants: genetic control of meiotic recombination and chromosome segregation, pp. 351–434 in The Genetics and Biology of Drosophila, Vol. 1a, edited by M. Ashburner and E. Novitski. Academic Press, New York.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

