Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder
- PMID: 17507910
- PMCID: PMC2629613
- DOI: 10.1038/sj.npp.1301457
Decreased mRNA expression of netrin-G1 and netrin-G2 in the temporal lobe in schizophrenia and bipolar disorder
Abstract
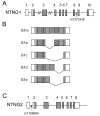
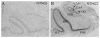
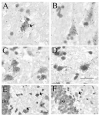
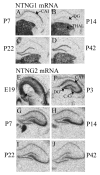
The membrane-bound axon guidance molecules netrin-G1 (NTNG1) and netrin-G2 (NTNG2) play a role in synaptic formation and maintenance. Non-coding single nucleotide polymorphisms (SNPs) in both genes have been reported to be associated with schizophrenia. The main aim of this study was to determine if NTNG1 and NTNG2 mRNA expression is altered in schizophrenia or bipolar disorder, and/or influenced by disease-associated SNPs. NTNG1 and NTNG2 mRNAs were examined in the medial and inferior temporal lobe using in situ hybridization and RT-PCR in the Stanley Medical Research Institute array collection, and in rat hippocampus during development and after antipsychotic administration. NTNG1 mRNA isoforms were also examined during human brain development. For NTNG1, the G1c isoform was reduced in bipolar disorder and with a similar trend in schizophrenia; expression of four other NTNG1 isoforms was unchanged. In both schizophrenia and bipolar disorder, NTNG2 mRNA was reduced in CA3, with reductions also found in CA4 and perirhinal cortex in bipolar disorder. The SNPs did not affect NTNG1 or NTNG2 mRNA expression. Both NTNG1 and NTNG2 mRNAs were developmentally regulated, and were unaltered by haloperidol, but NTNG2 mRNA was modestly increased by clozapine. These data implicate NTNG1 and NTNG2 in the pathophysiology of schizophrenia and bipolar disorder, but do not support the hypothesis that altered mRNA expression is the mechanism by which genetic variation of NTNG1 or NTNG2 may confer disease susceptibility.
Figures


Similar articles
-
A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia.Biol Psychiatry. 2005 Feb 15;57(4):382-93. doi: 10.1016/j.biopsych.2004.11.022. Biol Psychiatry. 2005. PMID: 15705354
-
Human netrin-G1 isoforms show evidence of differential expression.Genomics. 2005 Jul;86(1):112-6. doi: 10.1016/j.ygeno.2005.04.004. Genomics. 2005. PMID: 15901489
-
Vesicular glutamate transporter mRNA expression in the medial temporal lobe in major depressive disorder, bipolar disorder, and schizophrenia.Bipolar Disord. 2009 Nov;11(7):711-25. doi: 10.1111/j.1399-5618.2009.00752.x. Bipolar Disord. 2009. PMID: 19839996
-
Markers of glutamate synaptic transmission and plasticity are increased in the anterior cingulate cortex in bipolar disorder.Biol Psychiatry. 2010 Jun 1;67(11):1010-6. doi: 10.1016/j.biopsych.2009.12.004. Epub 2010 Jan 18. Biol Psychiatry. 2010. PMID: 20079890 Free PMC article.
-
Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia.Int J Dev Neurosci. 2011 May;29(3):311-24. doi: 10.1016/j.ijdevneu.2010.08.007. Epub 2010 Sep 15. Int J Dev Neurosci. 2011. PMID: 20833242 Free PMC article. Review.
Cited by
-
Immunoglobulin-Like Receptors and Their Impact on Wiring of Brain Synapses.Annu Rev Genet. 2018 Nov 23;52:567-590. doi: 10.1146/annurev-genet-120417-031513. Epub 2018 Sep 13. Annu Rev Genet. 2018. PMID: 30212237 Free PMC article. Review.
-
Identification of transcriptional regulatory elements for Ntng1 and Ntng2 genes in mice.Mol Brain. 2014 Mar 19;7:19. doi: 10.1186/1756-6606-7-19. Mol Brain. 2014. PMID: 24642214 Free PMC article.
-
FUS-mediated alternative splicing in the nervous system: consequences for ALS and FTLD.J Mol Med (Berl). 2013 Dec;91(12):1343-54. doi: 10.1007/s00109-013-1077-2. Epub 2013 Aug 24. J Mol Med (Berl). 2013. PMID: 23974990 Review.
-
Transcriptomic and neurochemical analysis of the stellate ganglia in mice highlights sex differences.Sci Rep. 2018 Jun 12;8(1):8963. doi: 10.1038/s41598-018-27306-3. Sci Rep. 2018. PMID: 29895973 Free PMC article.
-
Neurodevelopmental Perspectives on Wnt Signaling in Psychiatry.Mol Neuropsychiatry. 2017 Feb;2(4):219-246. doi: 10.1159/000453266. Epub 2017 Jan 13. Mol Neuropsychiatry. 2017. PMID: 28277568 Free PMC article. Review.
References
-
- Andreasen NC. A unitary model of schizophrenia: Bleuler's “fragmented phrene” as schizencephaly. Arch Gen Psychiatry. 1999;56:781–787. - PubMed
-
- Aoki-Suzuki M, Yamada K, Meerabux J, Iwayama-Shigeno Y, Ohba H, Iwamoto K, Takao H, Toyota T, Suto Y, Nakatani N, et al. A family-based association study and gene expression analyses of netrin-G1 and -G2 genes in schizophrenia. Biol Psychiatry. 2005;57:382–393. - PubMed
-
- Arnold SE. Cellular and molecular neuropathology of the parahippocampal region in schizophrenia. Ann NY Acad Sci. 2000;911:275–292. - PubMed
-
- Barton AJL, Najlerahim A, Pearson RCA, Harrison PJ. Pre- and postmortem influences on brain RNA. J Neurochem. 1993;61:1–11. - PubMed
-
- Biederer T. Hooking up new synapses. Nat Neurosci. 2006;9:1203–1204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

