Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers
- PMID: 17510386
- PMCID: PMC2858285
- DOI: 10.1158/0008-5472.CAN-06-3619
Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers
Abstract
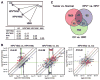
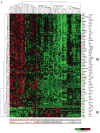
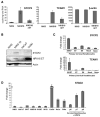
Human papillomaviruses (HPV) are associated with nearly all cervical cancers, 20% to 30% of head and neck cancers (HNC), and other cancers. Because HNCs also arise in HPV-negative patients, this type of cancer provides unique opportunities to define similarities and differences of HPV-positive versus HPV-negative cancers arising in the same tissue. Here, we describe genome-wide expression profiling of 84 HNCs, cervical cancers, and site-matched normal epithelial samples in which we used laser capture microdissection to enrich samples for tumor-derived versus normal epithelial cells. This analysis revealed that HPV(+) HNCs and cervical cancers differed in their patterns of gene expression yet shared many changes compared with HPV(-) HNCs. Some of these shared changes were predicted, but many others were not. Notably, HPV(+) HNCs and cervical cancers were found to be up-regulated in their expression of a distinct and larger subset of cell cycle genes than that observed in HPV(-) HNC. Moreover, HPV(+) cancers overexpressed testis-specific genes that are normally expressed only in meiotic cells. Many, although not all, of the hallmark differences between HPV(+) HNC and HPV(-) HNC were a direct consequence of HPV and in particular the viral E6 and E7 oncogenes. This included a novel association of HPV oncogenes with testis-specific gene expression. These findings in primary human tumors provide novel biomarkers for early detection of HPV(+) and HPV(-) cancers, and emphasize the potential value of targeting E6 and E7 function, alone or combined with radiation and/or traditional chemotherapy, in the treatment of HPV(+) cancers.
Figures



References
-
- Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363:1488–9. - PubMed
-
- Smith EM, Ritchie JM, Summersgill KF, et al. Human papillomavirus in oral exfoliated cells and risk of head and neck cancer. J Natl Cancer Inst. 2004;96:449–55. - PubMed
-
- Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nat Rev Cancer. 2005;5:127–35. - PubMed
-
- Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA101873/CA/NCI NIH HHS/United States
- DE15944/DE/NIDCR NIH HHS/United States
- R01 CA064364/CA/NCI NIH HHS/United States
- CA64364/CA/NCI NIH HHS/United States
- R29 CA064364/CA/NCI NIH HHS/United States
- R01 CA097944/CA/NCI NIH HHS/United States
- R01 CA078609/CA/NCI NIH HHS/United States
- CA078609/CA/NCI NIH HHS/United States
- R15 CA101873/CA/NCI NIH HHS/United States
- DE017315/DE/NIDCR NIH HHS/United States
- R01 DE017315/DE/NIDCR NIH HHS/United States
- CA100679/CA/NCI NIH HHS/United States
- CA97944/CA/NCI NIH HHS/United States
- R01 CA100679/CA/NCI NIH HHS/United States
- P01 CA022443/CA/NCI NIH HHS/United States
- R01 DE015944/DE/NIDCR NIH HHS/United States
- CA22443/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

