Predictive modeling of signaling crosstalk during C. elegans vulval development
- PMID: 17511512
- PMCID: PMC1868774
- DOI: 10.1371/journal.pcbi.0030092
Predictive modeling of signaling crosstalk during C. elegans vulval development
Abstract
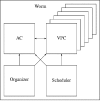
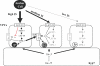
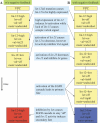
Caenorhabditis elegans vulval development provides an important paradigm for studying the process of cell fate determination and pattern formation during animal development. Although many genes controlling vulval cell fate specification have been identified, how they orchestrate themselves to generate a robust and invariant pattern of cell fates is not yet completely understood. Here, we have developed a dynamic computational model incorporating the current mechanistic understanding of gene interactions during this patterning process. A key feature of our model is the inclusion of multiple modes of crosstalk between the epidermal growth factor receptor (EGFR) and LIN-12/Notch signaling pathways, which together determine the fates of the six vulval precursor cells (VPCs). Computational analysis, using the model-checking technique, provides new biological insights into the regulatory network governing VPC fate specification and predicts novel negative feedback loops. In addition, our analysis shows that most mutations affecting vulval development lead to stable fate patterns in spite of variations in synchronicity between VPCs. Computational searches for the basis of this robustness show that a sequential activation of the EGFR-mediated inductive signaling and LIN-12 / Notch-mediated lateral signaling pathways is key to achieve a stable cell fate pattern. We demonstrate experimentally a time-delay between the activation of the inductive and lateral signaling pathways in wild-type animals and the loss of sequential signaling in mutants showing unstable fate patterns; thus, validating two key predictions provided by our modeling work. The insights gained by our modeling study further substantiate the usefulness of executing and analyzing mechanistic models to investigate complex biological behaviors.
Conflict of interest statement
Figures

References
-
- Fisher J, Henzinger TA. Executable biology. In Proceedings of the 39th Winter Simulation Conference; Track on Modeling and Simulation in Computational Biology; 3–6 December, 2006;; Monterey, California, United States.. Monterey (California): IEEE Computer Society Press; 2006. pp. 1675–1682.
-
- Fisher J, Harel D, Hubbard EJ, Piterman N, Stern MJ, et al. Combining state-based and scenario-based approaches in modeling biological systems. Lecture Notes Comp Sci; CMSB;; 26–28 May 2004;; Paris, France.. 2004. pp. 236–241. In. Proceedings of the 2nd International Conference on Computational Methods in Systems Biology.
-
- Clarke EM, Grumberg O, Peled D. Model checking. Cambridge (Massachusetts): The MIT Press; 1999. 330
-
- Kam N, Harel D, Kugler H, Marelly R, Pnueli A, et al. Formal modeling of C. elegans development: A scenario-based approach. Lecture Notes Comp Sci; CMSB;; 24–26 February 2003;; Roverto, Italy.. 2003. pp. 4–20. In. Proceedings of the First International Workshop on Computational Methods in Systems Biology.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

