Methodological considerations for gene expression profiling of human brain
- PMID: 17512057
- PMCID: PMC3835340
- DOI: 10.1016/j.jneumeth.2007.03.022
Methodological considerations for gene expression profiling of human brain
Abstract
Gene expression profiles of postmortem brain tissue represent important resources for understanding neuropsychiatric illnesses. The impact(s) of quality covariables on the analysis and results of gene expression studies are important questions. This paper addressed critical variables which might affect gene expression in two brain regions. Four broad groups of quality indicators in gene expression profiling studies (clinical, tissue, RNA, and microarray quality) were identified. These quality control indicators were significantly correlated, however one quality variable did not account for the total variance in microarray gene expression. The data showed that agonal factors and low pH correlated with decreased integrity of extracted RNA in two brain regions. These three parameters also modulated the significance of alterations in mitochondrial-related genes. The average F-ratio summaries across all transcripts showed that RNA degradation from the AffyRNAdeg program accounted for higher variation than all other quality factors. Taken together, these findings confirmed prior studies, which indicated that quality parameters including RNA integrity, agonal factors, and pH are related to differences in gene expression profiles in postmortem brain. Individual candidate genes can be evaluated with these quality parameters in post hoc analysis to help strengthen the relevance to psychiatric disorders. We find that clinical, tissue, RNA, and microarray quality are all useful variables for collection and consideration in study design, analysis, and interpretation of gene expression results in human postmortem studies.
Figures
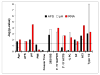
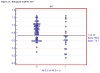
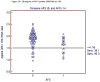

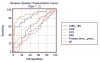
References
-
- Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58(2):85–96. - PubMed
-
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K, Marcucci U. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet. 2003;35(1):106. - PubMed
-
- Bahn S, Augood SJ, Ryan M, Standaert DG, Starkey M, Emson PC. Gene expression profiling in the post-mortem human brain--no cause for dismay. J Chem Neuroanat. 2001;22(1-2):79–94. - PubMed
-
- Barrett T, Cheadle C, Wood WB, Teichberg D, Donovan DM, Freed WJ, Becker KG, Vawter MP. Assembly and use of a broadly applicable neural cDNA microarray. Restor Neurol Neurosci. 2001;18(2-3):127–35. - PubMed
-
- Barton AJ, P R, Najlerahim A, Harrison PJ. Pre- and postmortem influences on brain RNA. J Neurochem. 1993;61(1):1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

