Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues
- PMID: 17512962
- PMCID: PMC2974173
- DOI: 10.1016/j.taap.2007.03.032
Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues
Abstract
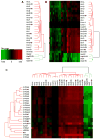
Frequently, primary hepatocytes are used as an in vitro model for the liver in vivo. However, the culture conditions reported vary considerably, with associated variability in performance. In this study, we characterized the differentiation character of primary human hepatocytes cultured using a highly defined, serum-free two-dimensional sandwich system, one that configures hepatocytes with collagen I as the substratum together with a dilute extracellular matrix (Matrigeltrade mark) overlay combined with a defined serum-free medium containing nanomolar levels of dexamethasone. Gap junctional communication, indicated by immunochemical detection of connexin 32 protein, was markedly enhanced in hepatocytes cultured in the Matrigel sandwich configuration. Whole genome expression profiling enabled direct comparison of liver tissues to hepatocytes and to the hepatoma-derived cell lines, HepG2 and Huh7. PANTHER database analyses were used to identify biological processes that were comparatively over-represented among probe sets expressed in the in vitro systems. The robustness of the primary hepatocyte cultures was reflected by the extent of unchanged expression character when compared directly to liver, with more than 77% of the probe sets unchanged in each of the over-represented categories, representing such genes as C/EBPalpha, HNF4alpha, CYP2D6, and ABCB1. In contrast, HepG2 and Huh7 cells were unchanged from the liver tissues for fewer than 48% and 55% of these probe sets, respectively. Further, hierarchical clustering of the hepatocytes, but not the cell lines, shifted from donor-specific to treatment-specific when the probe sets were filtered to focus on phenobarbital-inducible genes, indicative of the highly differentiated nature of the hepatocytes when cultured in a highly defined two-dimensional sandwich system.
Figures






References
-
- Annaert PP, Brouwer KL. Assessment of drug interactions in hepatobiliary transport using rhodamine 123 in sandwich-cultured rat hepatocytes. Drug Metab Dispos. 2005;33:388–394. - PubMed
-
- Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J Biol Chem. 2004;279:22250–22257. - PubMed
-
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. - PubMed
-
- Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, Suter L. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicol Sci. 2003;73:386–402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

