Network analysis of FDA approved drugs and their targets
- PMID: 17516560
- PMCID: PMC2561141
- DOI: 10.1002/msj.20002
Network analysis of FDA approved drugs and their targets
Abstract
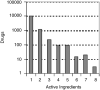
The global relationship between drugs that are approved for therapeutic use and the human genome is not known. We employed graph-theory methods to analyze the Federal Food and Drug Administration (FDA) approved drugs and their known molecular targets. We used the FDA Approved Drug Products with Therapeutic Equivalence Evaluations 26(th) Edition Electronic Orange Book (EOB) to identify all FDA approved drugs and their active ingredients. We then connected the list of active ingredients extracted from the EOB to those known human protein targets included in the DrugBank database and constructed a bipartite network. We computed network statistics and conducted Gene Ontology analysis on the drug targets and drug categories. We find that drug to drug-target relationship in the bipartite network is scale-free. Several classes of proteins in the human genome appear to be better targets for drugs since they appear to be selectively enriched as drug targets for the currently FDA approved drugs. These initial observations allow for development of an integrated research methodology to identify general principles of the drug discovery process.
Copyright (c) 2007 Mount Sinai School of Medicine.
Figures





References
-
- Albert R. Scale-free networks in cell biology. JCell Sci. 2005;118(Pt 21):4947–4957. - PubMed
-
- U.S. Food and Drug Administration. Center for Drug Evaluation and Research Approved Drug Products with Therapeutic Equivalence Evaluations Orange Book. http://www.fda.gov/cder/orange/default.htm.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

