Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7
- PMID: 17517131
- PMCID: PMC1891291
- DOI: 10.1186/1471-213X-7-53
Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7
Abstract
Background: Cdkn1c encodes an embryonic cyclin-dependant kinase inhibitor that acts to negatively regulate cell proliferation and, in some tissues, to actively direct differentiation. This gene, which is an imprinted gene expressed only from the maternal allele, lies within a complex region on mouse distal chromosome 7, called the IC2 domain, which contains several other imprinted genes. Studies on mouse embryos suggest a key role for genomic imprinting in regulating embryonic growth and this has led to the proposal that imprinting evolved as a consequence of the mismatched contribution of parental resources in mammals.
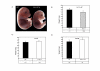
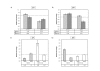
Results: In this study, we characterised the phenotype of mice carrying different copy number integrations of a bacterial artificial chromosome spanning Cdkn1c. Excess Cdkn1c resulted in embryonic growth retardation that was dosage-dependent and also responsive to the genetic background. Two-fold expression of Cdkn1c in a subset of tissues caused a 10-30% reduction in embryonic weight, embryonic lethality and was associated with a reduction in the expression of the potent, non-imprinted embryonic growth factor, Igf1. Conversely, loss of expression of Cdkn1c resulted in embryos that were 11% heavier with a two-fold increase in Igf1.
Conclusion: We have shown that embryonic growth in mice is exquisitely sensitive to the precise dosage of Cdkn1c. Cdkn1c is a maternally expressed gene and our findings support the prediction of the parental conflict hypothesis that that the paternal genome silences genes that have an inhibitory role in embryonic growth. Within the IC2 imprinted domain, Cdkn1c encodes the major regulator of embryonic growth and we propose that Cdkn1c was the focal point of the selective pressure for imprinting of this domain.
Figures


References
-
- Efstratiadis A. Genetics of mouse growth. Int J Dev Biol. 1998;42:955–976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

