Early sorting of inner nuclear membrane proteins is conserved
- PMID: 17517639
- PMCID: PMC1874229
- DOI: 10.1073/pnas.0703186104
Early sorting of inner nuclear membrane proteins is conserved
Abstract
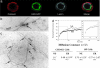
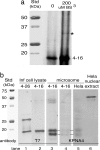
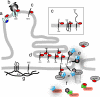
Spodoptera frugiperda (Sf9) importin-alpha-16 is a translocon-associated protein that participates in the early sorting pathway of baculovirus integral membrane proteins destined for the inner nuclear membrane (INM). To discern whether sorting intermediate protein complexes like those observed in insect cells are also formed with mammalian INM proteins, cross-linked complexes of importin-alpha-16 with human lamin B receptor (LBR) and nurim were examined. Both LBR and nurim cross-link with Sf9 importin-alpha-16 during cotranslational membrane integration and remain proximal with importin-alpha-16 after integration into the endoplasmic reticulum membrane and release from the translocon. Human cells encode several isoforms of importin-alpha; to determine whether any of these isoforms may recognize INM-directed proteins, they were tested for their ability to cross-link with the viral-derived INM sorting motif sequence. One cross-linked adduct was detected with a 16-kDa isoform encoded from KPNA4 (KPNA-4-16). KPNA-4-16 was easily detected in microsomal membranes prepared from KPNA4-16 recombinant virus-infected cells and was also detected in microsomes prepared from HeLa cells. Together these observations suggest that elements of the early sorting pathway of INM-directed proteins mediated by importin-alpha-16 are highly conserved, and mammalian KPNA-4-16 is a candidate partner in sorting integral membrane proteins to the INM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

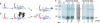
References
-
- Saksena S, Summers MD, Burks JK, Johnson AE, Braunagel SC. Nat Struct Mol Biol. 2006;13:500–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

