A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression
- PMID: 17517653
- PMCID: PMC1890503
- DOI: 10.1073/pnas.0610279104
A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression
Abstract
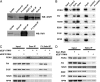
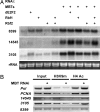
Regulation of chromatin structure is critical in many fundamental cellular processes. Previous studies have suggested that the Rb tumor suppressor may recruit multiple chromatin regulatory proteins to repress E2F, a key regulator of cell proliferation and differentiation. Taking advantage of the evolutionary conservation of the E2F pathway, we have conducted a genome-wide RNAi screen in cultured Drosophila cells for genes required for repression of E2F activity. Among the genes identified are components of the putative Domino chromatin remodeling complex, as well as the Polycomb Group (PcG) protein-like fly tumor suppressor, L3mbt, and the related CG16975/dSfmbt. These factors are recruited to E2F-responsive promoters through physical association with E2F and are required for repression of endogenous E2F target genes. Surprisingly, their inhibitory activities on E2F appear to be independent of Rb. In Drosophila, domino mutation enhances cell proliferation induced by E2F overexpression and suppresses a loss-of-function cyclin E mutation. These findings suggest that potential chromatin regulation mediated by Domino and PcG-like factors plays an important role in controlling E2F activity and cell growth.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lund AH, van Lohuizen M. Genes Dev. 2004;18:2315–2335. - PubMed
-
- Trimarchi JM, Lees JA. Nat Rev Mol Cell Biol. 2002;3:11–20. - PubMed
-
- Classon M, Harlow E. Nat Rev Cancer. 2002;2:910–917. - PubMed
-
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Nat Genet. 2005;37:579–583. - PubMed
-
- Frolov MV, Dyson NJ. J Cell Sci. 2004;117:2173–2181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

