Assisted large fragment insertion by Red/ET-recombination (ALFIRE)--an alternative and enhanced method for large fragment recombineering
- PMID: 17517785
- PMCID: PMC1904275
- DOI: 10.1093/nar/gkm250
Assisted large fragment insertion by Red/ET-recombination (ALFIRE)--an alternative and enhanced method for large fragment recombineering
Abstract
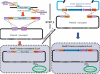
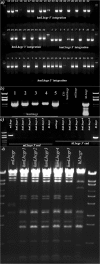
Functional genomics require manipulation and modification of large fragments of the genome. Such manipulation has only recently become more efficient due to the discovery of different techniques based on homologous recombination. However, certain limitations of these strategies still exist since insertion of homology arms (HAs) is often based on amplification of DNA sequences with PCR. Large quantities of PCR products longer than 4-5 kb can be difficult to obtain and the risk of mutations or mismatches increases with the size of the template that is being amplified. This can be overcome by adding HAs by conventional cloning techniques, but with large fragments such as entire genes the procedure becomes time-consuming and tedious. Second, homologous recombination techniques often require addition of antibiotic selection genes, which may not be desired in the final construct. Here, we report a method to overcome the size and selection marker limitations by a two- or three-step procedure. The method can insert any fragment into small or large episomes, without the need of an antibiotic selection gene. We have humanized the mouse luteinizing hormone receptor gene (Lhcgr) by inserting a approximately 55 kb fragment from a BAC clone containing the human Lhcgr gene into a 170 kb BAC clone comprising the entire mouse orthologue. The methodology is based on the rationale to introduce a counter-selection cassette flanked by unique restriction sites and HAs for the insert, into the vector that is modified. Upon enzymatic digestion, in vitro or in Escherichia coli, double-strand breaks are generated leading to recombination between the vector and the insert. The procedure described here is thus an additional powerful tool for manipulating large and complex genomic fragments.
Figures

References
-
- Sternberg NL. Cloning high molecular weight DNA fragments by the bacteriophage P1 system. Trends Genet. 1992;8:11–16. - PubMed
-
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat. Genet. 1994;6:84–89. - PubMed
-
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. - PubMed
-
- Muyrers JP, Zhang Y, Stewart AF. ET-cloning: think recombination first. Genet. Eng. (NY) 2000;22:77–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

