The NMR structure of the murine DLC2 SAM domain reveals a variant fold that is similar to a four-helix bundle
- PMID: 17519008
- PMCID: PMC1899508
- DOI: 10.1186/1472-6807-7-34
The NMR structure of the murine DLC2 SAM domain reveals a variant fold that is similar to a four-helix bundle
Abstract
Background: The tumor suppressor DLC2 (Deleted in Liver Cancer -2) participates in cell signaling at the mitochondrial membrane. DLC2 is characterized by a SAM (sterile alpha motif) domain, a Rho GTPase activating protein (GAP) domain, and a START lipid transfer domain.
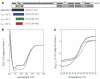
Results: Towards understanding the function of DLC2, we have solved the NMR solution structure of the SAM domain. The DLC2-SAM domain structure reveals an atypical four-helix composition that is distinct from the five-helix SAM domain structures that have been determined to date. From structural alignments, helix 3 of the canonical SAM domain appears to be replaced by shorter, extended secondary structure that follows a similar path. Another difference is demonstrated by helices 1 and 2 that form a helical hairpin that is situated approximately parallel to the canonical helix 5.
Conclusion: The DLC2-SAM domain adopts a structure that is topologically more similar to an anti-parallel four-helix bundle than a canonical SAM domain. This alternate topology may allow the DLC2-SAM domain to interact with a novel set of ligands.
Figures

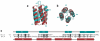

References
-
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. - PubMed
-
- Ching YP, Wong CM, Chan SF, Leung TH, Ng DC, Jin DY, Ng IO. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. The Journal of biological chemistry. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. - DOI - PubMed
-
- Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. International journal of oncology. 2006;29:1127–1132. - PubMed
-
- Ng IO, Liang ZD, Cao L, Lee TK. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

