The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation)
- PMID: 17521290
- PMCID: PMC2747773
- DOI: 10.1042/BJ20061890
The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation)
Abstract
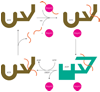
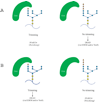
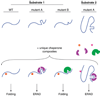
Over one-third of all newly synthesized polypeptides in eukaryotes interact with or insert into the membrane or the lumenal space of the ER (endoplasmic reticulum), an event that is essential for the subsequent folding, post-translational modification, assembly and targeting of these proteins. Consequently, the ER houses a large number of factors that catalyse protein maturation, but, in the event that maturation is aborted or inefficient, the resulting aberrant proteins may be selected for ERAD (ER-associated degradation). Many of the factors that augment protein biogenesis in the ER and that mediate ERAD substrate selection are molecular chaperones, some of which are heat- and/or stress-inducible and are thus known as Hsps (heat-shock proteins). But, regardless of whether they are constitutively expressed or are inducible, it has been assumed that all molecular chaperones function identically. As presented in this review, this assumption may be false. Instead, a growing body of evidence suggests that a chaperone might be involved in either folding or degrading a given substrate that transits through the ER. A deeper appreciation of this fact is critical because (i) the destruction of some ERAD substrates results in specific diseases, and (ii) altered ERAD efficiency might predispose individuals to metabolic disorders. Moreover, a growing number of chaperone-modulating drugs are being developed to treat maladies that arise from the synthesis of a unique mutant protein; therefore it is critical to understand how altering the activity of a single chaperone will affect the quality control of other nascent proteins that enter the ER.
Figures
References
-
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. - PubMed
-
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. - PubMed
-
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. - PubMed
-
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. - PubMed
-
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

