Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways
- PMID: 17521938
- PMCID: PMC1978064
- DOI: 10.1016/j.ymgme.2007.03.011
Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways
Abstract
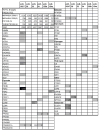
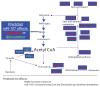
MicroRNAs (miRNAs) are powerful regulators of gene expression. Although first discovered in worm larvae, miRNAs play fundamental biological roles-including in humans-well beyond development. MiRNAs participate in the regulation of metabolism (including lipid metabolism) for all animal species studied. A review of the fascinating and fast-growing literature on miRNA regulation of metabolism can be parsed into three main categories: (1) adipocyte biochemistry and cell fate determination; (2) regulation of metabolic biochemistry in invertebrates; and (3) regulation of metabolic biochemistry in mammals. Most research into the 'function' of a given miRNA in metabolic pathways has concentrated on a given miRNA acting upon a particular 'target' mRNA. Whereas in some biological contexts the effects of a given miRNA:mRNA pair may predominate, this might not be the case generally. In order to provide an example of how a single miRNA could regulate multiple 'target' mRNAs or even entire human metabolic pathways, we include a discussion of metabolic pathways that are predicted to be regulated by the miRNA paralogs, miR-103 and miR-107. These miRNAs, which exist in vertebrate genomes within introns of the pantothenate kinase (PANK) genes, are predicted by bioinformatics to affect multiple mRNA targets in pathways that involve cellular Acetyl-CoA and lipid levels. Significantly, PANK enzymes also affect these pathways, so the miRNA and 'host' gene may act synergistically. These predictions require experimental verification. In conclusion, a review of the literature on miRNA regulation of metabolism leads us believe that the future will provide researchers with many additional energizing revelations.
Figures


References
-
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28(10):534–40. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. - PubMed
-
- Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–9. - PubMed
-
- Kosik KS, Krichevsky AM. The Elegance of the MicroRNAs: A Neuronal Perspective. Neuron. 2005;47(6):779–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

