CCR5Delta32 protein expression and stability are critical for resistance to human immunodeficiency virus type 1 in vivo
- PMID: 17522201
- PMCID: PMC1951285
- DOI: 10.1128/JVI.00068-07
CCR5Delta32 protein expression and stability are critical for resistance to human immunodeficiency virus type 1 in vivo
Abstract
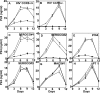
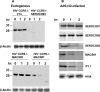
Human immunodeficiency virus type 1 (HIV-1) infection of individuals carrying the two alleles of the CCR5Delta32 mutation (CCR5(-/-)) has rarely been reported, but how the virus overcomes the CCR5Delta32 protective effect in these cases has not been delineated. We have investigated this in 6 infected (HIV(+)) and 25 HIV(-) CCR5(-/-) individuals. CD4(+) T lymphocytes isolated from HIV(-) CCR5(-/-) peripheral blood mononuclear cells (PBMCs) showed lower levels of CXCR4 expression that correlated with lower X4 Env-mediated fusion. Endogenous CCR5Delta32 protein was detected in all HIV(-) CCR5(-/-) PBMC samples (n = 25) but not in four of six unrelated HIV(+) CCR5(-/-) PBMC samples. Low levels were detected in another two HIV(+) CCR5(-/-) PBMC samples. The expression of adenovirus 5 (Ad5)-encoded CCR5Delta32 protein restored the protective effect in PBMCs from three HIV(+) CCR5(-/-) individuals but failed to restore the protective effect in PBMCs isolated from another three HIV(+) CCR5(-/-) individuals. In the latter samples, pulse-chase analyses demonstrated the disappearance of endogenous Ad5-encoded CCR5Delta32 protein and the accumulation of Ad5-encoded CCR5 during the chase periods. PBMCs isolated from CCR5(-/-) individuals showed resistance to primary X4 but were readily infected by a lab-adapted X4 strain. Low levels of Ad5-encoded CCR5Delta32 protein conferred resistance to primary X4 but not to lab-adapted X4 virus. These data provide strong support for the hypothesis that the CCR5Delta32 protein actively confers resistance to HIV-1 in vivo and suggest that the loss or reduction of CCR5Delta32 protein expression may account for HIV-1 infection of CCR5(-/-) individuals. The results also suggest that other cellular or virally induced factors may be involved in the stability of CCR5Delta32 protein.
Figures


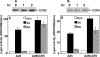


References
-
- Agrawal, L., X. Lu, Q. Jin, and G. Alkhatib. 2006. Anti-HIV therapy: current and future directions. Curr. Pharm. Des. 12:2031-2055. - PubMed
-
- Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. - PubMed
-
- Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

