Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant
- PMID: 17522211
- PMCID: PMC1951302
- DOI: 10.1128/JVI.02694-06
Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant
Abstract
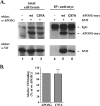
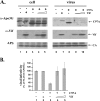
Human immunodeficiency virus type 1 (HIV-1) Vif counteracts the antiviral activity of the human cytidine deaminase APOBEC3G (APO3G) by inhibiting its incorporation into virions. This has been attributed to the Vif-induced degradation of APO3G by cytoplasmic proteasomes. We recently demonstrated that although APO3G has a natural tendency to form RNA-dependent homo-multimers, multimerization was not essential for encapsidation into HIV-1 virions or antiviral activity. We now demonstrate that a multimerization-defective APO3G variant (APO3G C97A) is able to assemble into RNase-sensitive high-molecular-mass (HMM) complexes, suggesting that homo-multimerization of APO3G and assembly into HMM complexes are unrelated RNA-dependent processes. Interestingly, APO3G C97A was highly resistant to Vif-induced degradation even though the two proteins were found to interact in coimmunoprecipitation experiments and exhibited partial colocalization in transfected HeLa cells. Surprisingly, encapsidation and antiviral activity of APO3G C97A were both inhibited by Vif despite resistance to degradation. These results demonstrate that targeting of APO3G to proteasome degradation and interference with viral encapsidation are distinct functional properties of Vif.
Figures





References
-
- Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. - PubMed
-
- Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

