Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F
- PMID: 17522216
- PMCID: PMC1951317
- DOI: 10.1128/JVI.00395-07
Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F
Abstract
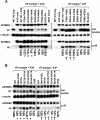
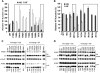
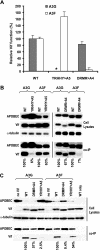
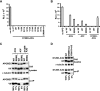
Human cytidine deaminases APOBEC3G (A3G) and APOBEC3F (A3F) inhibit replication of Vif-deficient human immunodeficiency virus type 1 (HIV-1). HIV-1 Vif overcomes these host restriction factors by binding to them and inducing their proteasomal degradation. The Vif-A3G and Vif-A3F interactions are attractive targets for antiviral drug development because inhibiting the interactions could allow the host defense mechanism to control HIV-1 replication. It was recently reported that the Vif amino acids D(14)RMR(17) are important for functional interaction and degradation of the previously identified Vif-resistant mutant of A3G (D128K-A3G). However, the Vif determinants important for functional interaction with A3G and A3F have not been fully characterized. To identify these determinants, we performed an extensive mutational analysis of HIV-1 Vif. Our analysis revealed two distinct Vif determinants, amino acids Y(40)RHHY(44) and D(14)RMR(17), which are essential for binding to A3G and A3F, respectively. Interestingly, mutation of the A3G-binding region increased Vif's ability to suppress A3F. Vif binding to D128K-A3G was also dependent on the Y(40)RHHY(44) region but not the D(14)RMR(17) region. Consistent with previous observations, subsequent neutralization of the D128K-A3G antiviral activity required substitution of Vif determinant D(14)RMR(17) with SEMQ, similar to the SERQ amino acids in simian immunodeficiency virus SIV(AGM) Vif, which is capable of neutralizing D128K-A3G. These studies are the first to clearly identify two distinct regions of Vif that are critical for independent interactions with A3G and A3F. Pharmacological interference with the Vif-A3G or Vif-A3F interactions could result in potent inhibition of HIV-1 replication by the APOBEC3 proteins.
Figures
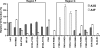
References
-
- Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. - PubMed
-
- Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. - PMC - PubMed
-
- Douaisi, M., S. Dussart, M. Courcoul, G. Bessou, E. C. Lerner, E. Decroly, and R. Vigne. 2005. The tyrosine kinases Fyn and Hck favor the recruitment of tyrosine-phosphorylated APOBEC3G into vif-defective HIV-1 particles. Biochem. Biophys. Res. Commun. 329:917-924. - PubMed
-
- Fujita, M., A. Sakurai, A. Yoshida, M. Miyaura, A. H. Koyama, K. Sakai, and A. Adachi. 2003. Amino acid residues 88 and 89 in the central hydrophilic region of human immunodeficiency virus type 1 Vif are critical for viral infectivity by enhancing the steady-state expression of Vif. J. Virol. 77:1626-1632. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

