The NS3 helicase and NS5B-to-3'X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells
- PMID: 17522229
- PMCID: PMC1951293
- DOI: 10.1128/JVI.02088-06
The NS3 helicase and NS5B-to-3'X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells
Abstract
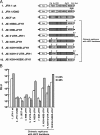
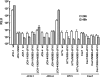
The JFH-1 strain of hepatitis C virus (HCV) is a genotype 2a strain that can replicate autonomously in Huh7 cells. The J6 strain is also a genotype 2a strain, but its full genomic RNA does not replicate in Huh7 cells. However, chimeric J6/JFH-1 RNA that has J6 structural-protein-coding regions and JFH-1 nonstructural-protein-coding regions can replicate autonomously and produce infectious HCV particles. In order to determine the mechanisms underlying JFH-1 RNA replication, we constructed various J6/JFH-1 chimeras and tested their RNA replication and virus particle production abilities in Huh7 cells. Via subgenomic-RNA-replication assays, we found that both the JFH-1 NS5B-to-3'X (N5BX) and the NS3 helicase (N3H) regions are important for the replication of the J6CF replicon. We applied these results to full-length genomic RNA replication and analyzed replication using Northern blotting. We found that a chimeric J6 clone with JFH-1 N3H and N5BX could replicate autonomously but that a chimeric J6 clone with only JFH-1 N5BX had no replication ability. Finally, we tested the virus production abilities of these clones and found that a chimeric J6 clone with JFH-1 N3H and N5BX could produce infectious HCV particles. In conclusion, the JFH-1 NS3 helicase and NS5B-to-3'X regions are important for efficient replication and virus particle formation of HCV genotype 2a strains.
Figures



References
-
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
-
- Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. - PubMed
-
- Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

