A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples
- PMID: 17524135
- PMCID: PMC1888694
- DOI: 10.1186/1471-2180-7-47
A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples
Abstract
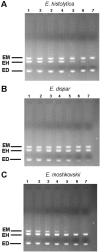
Background: E. histolytica, a pathogenic amoeba, is indistinguishable in its cyst and trophozoite stages from those of non-pathogenic E. moshkovskii and E. dispar by light microscopy. We have developed a nested multiplex PCR targeting a 16S-like rRNA gene for differential detection of all the three morphologically similar forms of E. histolytica, E. moshkovskii and E. dispar simultaneously in stool samples.
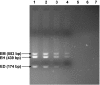
Results: The species specific product size for E. histolytica, E. moshkovskii and E. dispar was 439, 553 and 174 bp respectively, which was clearly different for all the three Entamoeba species. The nested multiplex PCR showed a sensitivity of 94% and specificity of 100% for the demonstration of E. histolytica, E. moshkovskii and E. dispar DNA in stool samples. The PCR was positive for E. histolytica, E. moshkovskii and E. dispar in a total of 190 out of 202 stool specimens (94% sensitive) that were positive for E. histolytica/E. dispar/E. moshkovskii by examination of stool by microscopy and/or culture. All the 35 negative control stool samples that were negative for E. histolytica/E. dispar/E. moshkovskii by microscopy and culture were also found negative by the nested multiplex PCR (100% specific). The result from the study shows that only 34.6% of the patient stool samples that were positive for E. histolytica/E. dispar/E. moshkovskii by examination of stool by microscopy and/or culture, were actually positive for pathogenic E. histolytica and the remaining majority of the stool samples were positive for non-pathogenic E. dispar or E. moshkovskii as demonstrated by the use of nested multiplex PCR.
Conclusion: The present study reports a new nested multiplex PCR strategy for species specific detection and differentiation of E. histolytica, E. dispar and E. moshkovskii DNA in stool specimens. The test is highly specific, sensitive and also rapid, providing the results within 12 hours of receiving stool specimens.
Figures
References
-
- World Health Organization Amoebiasis. WHO Weekly Epidemiol Rec. 1997;72:97–100.
-
- Krogstad DJ, Spencer HC, Jr, Healy GR, Gleason NN, Sexton DJ, Herron CA. Amebiasis: epidemiologic studies in the United States, 1971–1974. Ann Intern Med. 1978;88:89–97. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

