Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase
- PMID: 17524144
- PMCID: PMC1904441
- DOI: 10.1186/1743-422X-4-44
Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase
Abstract
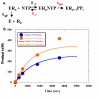
Background: Putative high mutation rates of RNA viruses are believed to mediate undesirable phenomena, such as emergence of drug resistance. However, very little is known about biochemical fidelity rates for viral RNA-dependent RNA polymerases. Using a recently developed in vitro polymerase assay for poliovirus polymerase 3Dpol [Arnold and Cameron (2000) JBC 275:5329], we measured fidelity for each possible mismatch. Polymerase fidelity, in contrast to sequence error rate, is biochemically defined as kpol/Kd of {(correct plus incorrect) divided by incorrect} incorporations, such that a larger value connotes higher fidelity.
Results: To derive kpol/Kd for correct base incorporation, we performed conventional pre-steady state single turnover measurements, yielding values that range from 0.62 to 9.4 microM-1 sec-1. Pre-steady state measurements for incorrect base incorporation were less straightforward: several anomalous phenomena interfered with data collection. To obtain pre-steady state kinetic data for incorrect base incorporation, three strategies were employed. (1) For some incorrect bases, a conventional approach was feasible, although care was taken to ensure that only single turnovers were being assessed. (2) Heparin or unlabeled RNA traps were used to simulate single turnover conditions. (3) Finally, for some incorrect bases, incorporation was so poor that single datapoints were used to provide kinetic estimates. Overall, we found that fidelity for poliovirus polymerase 3Dpol ranges from 1.2 x 10(4) to 1.0 x 10(6) for transition mutations and 3.2 x 10(5) to 4.3 x 10(7) for transversion mutations.
Conclusion: These values are unexpectedly high showing that high RNA virus sequence variation is not due to intrinsically low polymerase fidelity. Based on unusual enzyme behavior that we observed, we speculate that RNA mismatches either directly or indirectly cause enzyme RNA dissociation. If so, high sequence variation of RNA viruses may be due to template-switch RNA recombination and/or unknown fitness/selection phenomena. These findings may lead to a mechanistic understanding of RNA virus error catastrophe and improved anti-viral strategies.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

