Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey
- PMID: 17524145
- PMCID: PMC1894642
- DOI: 10.1186/1471-2164-8-132
Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey
Abstract
Background: Extensive computational and database tools are available to mine genomic and genetic databases for model organisms, but little genomic data is available for many species of ecological or agricultural significance, especially those with large genomes. Genome surveys using conventional sequencing techniques are powerful, particularly for detecting sequences present in many copies per genome. However these methods are time-consuming and have potential drawbacks. High throughput 454 sequencing provides an alternative method by which much information can be gained quickly and cheaply from high-coverage surveys of genomic DNA.
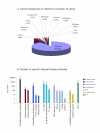
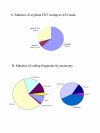
Results: We sequenced 78 million base-pairs of randomly sheared soybean DNA which passed our quality criteria. Computational analysis of the survey sequences provided global information on the abundant repetitive sequences in soybean. The sequence was used to determine the copy number across regions of large genomic clones or contigs and discover higher-order structures within satellite repeats. We have created an annotated, online database of sequences present in multiple copies in the soybean genome. The low bias of pyrosequencing against repeat sequences is demonstrated by the overall composition of the survey data, which matches well with past estimates of repetitive DNA content obtained by DNA re-association kinetics (Cot analysis).
Conclusion: This approach provides a potential aid to conventional or shotgun genome assembly, by allowing rapid assessment of copy number in any clone or clone-end sequence. In addition, we show that partial sequencing can provide access to partial protein-coding sequences.
Figures

References
-
- Fleishmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J-F, Dougherty BA, Merrick JM, McKenney K, Sutton G, FitzHugh W, Fields C, Gocyne JD, Scott J, Shirley R, Liu L-I, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotton MD, Utterback TR, Hanna MC, Nguyen DT, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

