The origin and virulence of the 1918 "Spanish" influenza virus
- PMID: 17526158
- PMCID: PMC2720273
The origin and virulence of the 1918 "Spanish" influenza virus
Abstract
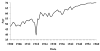
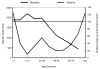
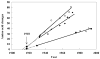
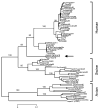
The "Spanish" influenza pandemic of 1918-19 caused acute illness in 25-30 percent of the world's population and resulted in the death of up to an estimated 40 million people. Using fixed and frozen lung tissue of 1918 influenza victims, the complete genomic sequence of the 1918 influenza virus has been deduced. Sequence and phylogenetic analysis of the completed 1918 influenza virus genes shows them to be the most avian-like among the mammalian-adapted viruses. This finding supports the hypotheses that (1) the pandemic virus contains genes derived from avian-like influenza virus strains and that (2) the 1918 virus is the common ancestor of human and classical swine H1N1 influenza viruses. The relationship of the 1918 virus with avian influenza viruses is further supported by recent work in which the 1918 hemagglutinin (HA) protein crystal structure was resolved. Neither the 1918 hemagglutinin (HA) nor the neuraminidase (NA) genes possess mutations known to increase tissue tropicity that account for the virulence of other influenza virus strains like A/WSN/33 or the highly pathogenic avian influenza H5 or H7 viruses. Using reverse genetics approaches, influenza virus constructs containing the 1918 HA and NA on a modern human influenza virus background were lethal in mice. The complete 1918 virus was even more virulent in mice. The genotypic basis of this virulence has not yet been elucidated. The complete sequence of the non-structural (NS) gene segment of the 1918 virus was deduced and also tested for the hypothesis that enhanced virulence in 1918 could have been due to type I interferon inhibition by the NS1 protein. Results from these experiments suggest that in human cells the 1918 NS1 is a very effective interferon antagonist, but the 1918 NS1 gene does not have the amino acid change that correlates with virulence in the H5N1 virus strains identified in 1997 in Hong Kong. Sequence analysis of the 1918 pandemic influenza virus is allowing us to test hypotheses as to the origin and virulence of this strain. This information should help elucidate how pandemic influenza virus strains emerge and what genetic features contribute to virulence in humans.
Figures

References
-
- Barry JM. The great influenza: The epic story of the deadliest plague in history. New York: Viking Press; 2004. p. 560.
-
- Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, García-Sastre A, Palese P, Taubenberger JK. Sequence of the 1918 pandemic influenza virus non-structural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci U S A. 2001;98:2746–51. - PMC - PubMed
-
- Beveridge W. Influenza: The last great plague, an unfinished story of discovery. New York: Prodist; 1977.
-
- Brown IH, Chakraverty P, Harris PA, Alexander DJ. Disease outbreaks in pigs in Great Britain due to an influenza A virus of H1N2 subtype. Vet Rec. 1995;136:328–29. - PubMed
-
- Brown IH, Harris PA, McCauley JW, Alexander DJ. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79:2947–55. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
