Functional sequestration of transcription factor activity by repetitive DNA
- PMID: 17526489
- PMCID: PMC3812952
- DOI: 10.1074/jbc.M702547200
Functional sequestration of transcription factor activity by repetitive DNA
Abstract
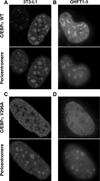
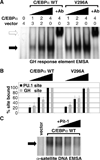
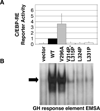
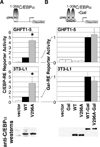
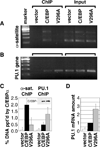
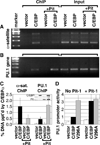
Higher eukaryote genomes contain repetitive DNAs, often concentrated in transcriptionally inactive heterochromatin. Although repetitive DNAs are not typically considered as regulatory elements that directly affect transcription, they can contain binding sites for some transcription factors. Here, we demonstrate that binding of the transcription factor CCAAT/enhancer-binding protein alpha (C/EBPalpha) to the mouse major alpha-satellite repetitive DNA sequesters C/EBPalpha in the transcriptionally inert pericentromeric heterochromatin. We find that this sequestration reduces the transcriptional capacity of C/EBPalpha. Functional sequestration of C/EBPalpha was demonstrated by experimentally reducing C/EBPalpha binding to the major alpha-satellite DNA, which elevated the concentration of C/EBPalpha in the non-heterochromatic subcompartment of the cell nucleus. The reduction in C/EBPalpha binding to alpha-satellite DNA was induced by the co-expression of the transcription factor Pit-1, which removes C/EBPalpha from the heterochromatic compartment, and by the introduction of an altered-specificity mutation into C/EBPalpha that reduces binding to alpha-satellite DNA but permits normal binding to sites in some gene promoters. In both cases the loss of alpha-satellite DNA binding coincided with an elevation in the binding of C/EBPalpha to a promoter and an increased transcriptional output from that promoter. Thus, the binding of C/EBPalpha to this highly repetitive DNA reduced the amount of C/EBPalpha available for binding to and regulation of this promoter. The functional sequestration of some transcription factors through binding to repetitive DNAs may represent an underappreciated mechanism controlling transcription output.
Figures

Similar articles
-
A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAAT/enhancer binding protein alpha required for prolactin gene transcription.Mol Endocrinol. 2003 Feb;17(2):209-22. doi: 10.1210/me.2001-0222. Mol Endocrinol. 2003. PMID: 12554749 Free PMC article.
-
TCERG1 inhibits C/EBPα through a mechanism that does not involve sequestration of C/EBPα at pericentromeric heterochromatin.J Cell Biochem. 2011 Sep;112(9):2317-26. doi: 10.1002/jcb.23154. J Cell Biochem. 2011. PMID: 21503969
-
Imaging the localized protein interactions between Pit-1 and the CCAAT/enhancer binding protein alpha in the living pituitary cell nucleus.Mol Endocrinol. 2003 Mar;17(3):333-45. doi: 10.1210/me.2002-0136. Epub 2002 Dec 12. Mol Endocrinol. 2003. PMID: 12554785 Free PMC article.
-
Satellite DNA-mediated effects on genome regulation.Genome Dyn. 2012;7:153-69. doi: 10.1159/000337116. Epub 2012 Jun 25. Genome Dyn. 2012. PMID: 22759818 Review.
-
Regulation of granulocyte and monocyte differentiation by CCAAT/enhancer binding protein alpha.Blood Cells Mol Dis. 2003 Nov-Dec;31(3):338-41. doi: 10.1016/s1079-9796(03)00135-9. Blood Cells Mol Dis. 2003. PMID: 14636649 Review.
Cited by
-
Indirect Mechanisms of Transcription Factor-Mediated Gene Regulation during Cell Fate Changes.Adv Genet (Hoboken). 2022 Nov 9;3(4):2200015. doi: 10.1002/ggn2.202200015. eCollection 2022 Dec. Adv Genet (Hoboken). 2022. PMID: 36911290 Free PMC article.
-
Structural basis of DNA methylation-dependent site selectivity of the Epstein-Barr virus lytic switch protein ZEBRA/Zta/BZLF1.Nucleic Acids Res. 2022 Jan 11;50(1):490-511. doi: 10.1093/nar/gkab1183. Nucleic Acids Res. 2022. PMID: 34893887 Free PMC article.
-
ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation.Autoimmunity. 2008 May;41(4):253-71. doi: 10.1080/08916930802024202. Autoimmunity. 2008. PMID: 18432406 Free PMC article. Review.
-
Evolution acts on enhancer organization to fine-tune gradient threshold readouts.PLoS Biol. 2008 Nov 4;6(11):e263. doi: 10.1371/journal.pbio.0060263. PLoS Biol. 2008. PMID: 18986212 Free PMC article.
-
Programmable gene regulation for metabolic engineering using decoy transcription factor binding sites.Nucleic Acids Res. 2021 Jan 25;49(2):1163-1172. doi: 10.1093/nar/gkaa1234. Nucleic Acids Res. 2021. PMID: 33367820 Free PMC article.
References
-
- Isogai Y, Tjian R. Curr Opin Cell Biol. 2003;15:1–8. - PubMed
-
- Kosak ST, Groudine M. Genes Dev. 2004;18:1371–1384. - PubMed
-
- Misteli T. Cell. 2004;119:153–156. - PubMed
-
- Henikoff S, Ahmad K. Ann Rev Cell Dev Biol. 2005;21:133–153. - PubMed
-
- Henikoff S, Ahmad K, Malik HS. Science. 2001;293:1098–1102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

