Conformational dynamics in loop swap mutants of homologous fibronectin type III domains
- PMID: 17526562
- PMCID: PMC1965443
- DOI: 10.1529/biophysj.106.100578
Conformational dynamics in loop swap mutants of homologous fibronectin type III domains
Abstract
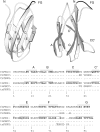
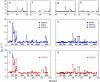
Fibronectin type III (FN-III) domains are autonomously folded modules found in a variety of multidomain proteins. The 10th FN-III domain from fibronectin (fnFN10) and the 3rd FN-III domain from tenascin-C (tnFN3) have 27% sequence identity and the same overall fold; however, the CC' loop has a different pattern of backbone hydrogen bonds and the FG loop is longer in fnFN10 compared to tnFN3. To examine the influence of length, sequence, and context in determining dynamical properties of loops, CC' and FG loops were swapped between fnFN10 and tnFN3 to generate four mutant proteins and backbone conformational dynamics on ps-ns and mus-ms timescales were characterized by solution (15)N-NMR spin relaxation spectroscopy. The grafted loops do not strongly perturb the properties of the protein scaffold; however, specific effects of the mutations are observed for amino acids that are proximal in space to the sites of mutation. The amino acid sequence primarily dictates conformational dynamics when the wild-type and grafted loop have the same length, but both sequence and context contribute to conformational dynamics when the loop lengths differ. The results suggest that changes in conformational dynamics of mutant proteins must be considered in both theoretical studies and protein design efforts.
Figures



Similar articles
-
Crosstalk between the protein surface and hydrophobic core in a core-swapped fibronectin type III domain.J Mol Biol. 2008 Jan 11;375(2):560-71. doi: 10.1016/j.jmb.2007.10.056. Epub 2007 Oct 25. J Mol Biol. 2008. PMID: 18035373 Free PMC article.
-
Backbone dynamics of homologous fibronectin type III cell adhesion domains from fibronectin and tenascin.Structure. 1997 Jul 15;5(7):949-59. doi: 10.1016/S0969-2126(97)00248-7. Structure. 1997. PMID: 9261088
-
Stabilization of the third fibronectin type III domain of human tenascin-C through minimal mutation and rational design.Protein Eng Des Sel. 2014 Oct;27(10):411-8. doi: 10.1093/protein/gzu024. Epub 2014 Jul 4. Protein Eng Des Sel. 2014. PMID: 24996411
-
Adnectins: engineered target-binding protein therapeutics.Protein Eng Des Sel. 2011 Jan;24(1-2):3-9. doi: 10.1093/protein/gzq097. Epub 2010 Nov 10. Protein Eng Des Sel. 2011. PMID: 21068165 Free PMC article. Review.
-
Building proteins with fibronectin type III modules.Structure. 1994 May 15;2(5):333-7. doi: 10.1016/s0969-2126(00)00034-4. Structure. 1994. PMID: 8081748 Review.
Cited by
-
AMP-activated protein kinase β-subunit requires internal motion for optimal carbohydrate binding.Biophys J. 2012 Jan 18;102(2):305-14. doi: 10.1016/j.bpj.2011.12.012. Biophys J. 2012. PMID: 22339867 Free PMC article.
-
Crosstalk between the protein surface and hydrophobic core in a core-swapped fibronectin type III domain.J Mol Biol. 2008 Jan 11;375(2):560-71. doi: 10.1016/j.jmb.2007.10.056. Epub 2007 Oct 25. J Mol Biol. 2008. PMID: 18035373 Free PMC article.
-
The flexibility of a distant loop modulates active site motion and product release in ribonuclease A.Biochemistry. 2009 Aug 4;48(30):7160-8. doi: 10.1021/bi900830g. Biochemistry. 2009. PMID: 19588901 Free PMC article.
-
Transient opening of fibronectin type III (FNIII) domains: the interaction of the third FNIII domain of FN with anastellin.Biochemistry. 2009 May 19;48(19):4189-97. doi: 10.1021/bi900001g. Biochemistry. 2009. PMID: 19320499 Free PMC article.
-
Structure and dynamics of Ca2+-binding domain 1 of the Na+/Ca2+ exchanger in the presence and in the absence of Ca2+.J Mol Biol. 2008 Mar 28;377(3):945-55. doi: 10.1016/j.jmb.2008.01.046. Epub 2008 Jan 30. J Mol Biol. 2008. PMID: 18280495 Free PMC article.
References
-
- James, L. C., P. Roversi, and D. S. Tawfik. 2003. Antibody multispecificity mediated by conformational diversity. Science. 299:1362–1367. - PubMed
-
- Boyd, A. E., C. S. Dunlop, L. Wong, Z. Radic, P. Taylor, and D. A. Johnson. 2004. Nanosecond dynamics of acetylcholinesterase near the active center gorge. J. Biol. Chem. 279:26612–26618. - PubMed
-
- Petrey, D., and B. Honig. 2005. Protein structure prediction: inroads to biology. Mol. Cell. 20:811–819. - PubMed
-
- Binz, H. K., and A. Pluckthun. 2005. Engineered proteins as specific binding reagents. Curr. Opin. Biotechnol. 16:459–469. - PubMed
-
- Palmer, A. G. 2004. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 104:3623–3640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

