Models of beta-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process
- PMID: 17526580
- PMCID: PMC1959551
- DOI: 10.1529/biophysj.107.110148
Models of beta-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process
Abstract
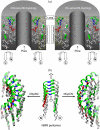
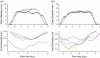
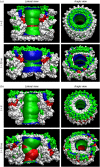
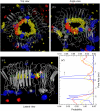
Here we model the Alzheimer beta-peptide ion channel with the goal of obtaining insight into the mechanism of amyloid toxicity. The models are built based on NMR data of the oligomers, with the universal U-shaped (strand-turn-strand) motif. After 30-ns simulations in the bilayer, the channel dimensions, shapes and subunit organization are in good agreement with atomic force microscopy (AFM). The models use the Abeta(17-42) pentamer NMR-based coordinates. Extension and bending of the straight oligomers lead to two channel topologies, depending on the direction of the curvature: 1), the polar/charged N-terminal beta-strand of Abeta(17-42) faces the water-filled pore, and the hydrophobic C-terminal beta-strand faces the bilayer (CNpNC; p for pore); and 2), the C-terminal beta-strand faces the solvated pore (NCpCN). In the atomistic simulations in a fully solvated DOPC lipid bilayer, the first (CNpNC) channel preserves the pore and conducts solvent; by contrast, hydrophobic collapse blocks the NCpCN channel. AFM demonstrated open pores and collapsed complexes. The final averaged CNpNC pore dimensions (outer diameter 8 nm; inner diameter approximately 2.5 nm) are in the AFM range (8-12 nm; approximately 2 nm, respectively). Further, in agreement with high-resolution AFM images, during the simulations, the channels spontaneously break into ordered subunits in the bilayer; however, we also observe that the subunits are loosely connected by partially disordered inner beta-sheet, suggesting subunit mobility in the bilayer. The cationic channel has strong selective affinity for Ca(2+), supporting experimental calcium-selective beta-amyloid channels. Membrane permeability and consequent disruption of calcium homeostasis were implicated in cellular degeneration. Consequently, the CNpNC channel topology can sign cell death, offering insight into amyloid toxicity via an ion "trap-release" transport mechanism. The observed loosely connected subunit organization suggests that amyloid channel formation in the bilayer is a dynamic, fluid process involving subunit association, dissociation, and channel rearrangements.
Figures




References
-
- Selkoe, D. J. 1991. Alzheimer's disease. In the beginning. Nature. 354:432–433. - PubMed
-
- Kirkitadze, M. D., G. Bitan, and D. B. Teplow. 2002. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 69:567–577. - PubMed
-
- Bucciantini, M., E. Giannoni, F. Chiti, F. Baroni, L. Formigli, J. Zurdo, N. Taddei, G. Ramponi, C. M. Dobson, and M. Stefani. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 416:507–511. - PubMed
-
- Walsh, D. M., I. Klyubin, J. V. Fadeeva, W. K. Cullen, R. Anwyl, M. S. Wolfe, M. J. Rowan, and D. J. Selkoe. 2002. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 416:535–539. - PubMed
-
- Lin, H., Y. J. Zhu, and R. Lal. 1999. Amyloid β protein (1–40) forms calcium-permeable, Zn2+-sensitive channel in reconstituted lipid vesicles. Biochemistry. 38:11189–11196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

