Nanometer distance measurements in RNA using site-directed spin labeling
- PMID: 17526583
- PMCID: PMC1959523
- DOI: 10.1529/biophysj.107.109439
Nanometer distance measurements in RNA using site-directed spin labeling
Abstract
The method of site-directed spin labeling (SDSL) utilizes a stable nitroxide radical to obtain structural and dynamic information on biomolecules. Measuring dipolar interactions between pairs of nitroxides yields internitroxide distances, from which quantitative structural information can be derived. This study evaluates SDSL distance measurements in RNA using a nitroxide probe, designated as R5, which is attached in an efficient and cost-effective manner to backbone phosphorothioate sites that are chemically substituted in arbitrary sequences. It is shown that R5 does not perturb the global structure of the A-form RNA helix. Six sets of internitroxide distances, ranging from 20 to 50 A, were measured on an RNA duplex with a known X-ray crystal structure. The measured distances strongly correlate (R(2) = 0.97) with those predicted using an efficient algorithm for determining the expected internitroxide distances from the parent RNA structure. The results enable future studies of global RNA structures for which high-resolution structural data are absent.
Figures



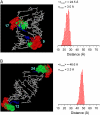

References
-
- Gesteland, R. F., J. F. Atkins, and T. R. Cech, editors. 2006. RNA World, 3rd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
-
- Moore, P. B., and T. A. Steitz. 2005. The ribosome revealed. Trends Biochem. Sci. 30:281–283. - PubMed
-
- Hubbell, W. L., A. Gross, R. Langen, and M. A. Leitzow. 1998. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 8:649–656. - PubMed
-
- Feix, J. B., and C. S. Klug. 1998. Site-directed spin labeling of membrane proteins and peptide-membrane interactions. In Biological Magnetic Resonance. L. J. Berliner, editor. Plenum Press, New York. 251–281.
-
- Hubbell, W. L., D. S. Cafiso, and C. Altenbach. 2000. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7:735–739. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

