Functional characterization of the Sinorhizobium meliloti acetate metabolism genes aceA, SMc00767, and glcB
- PMID: 17526694
- PMCID: PMC1952029
- DOI: 10.1128/JB.00385-07
Functional characterization of the Sinorhizobium meliloti acetate metabolism genes aceA, SMc00767, and glcB
Abstract
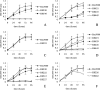
The genes encoding malate synthase (glcB) and isocitrate lyase (aceA) and a 240-bp open reading frame (SMc00767) located downstream of aceA were isolated and functionally characterized in Sinorhizobium meliloti. Independent and double interposon mutants of each gene were constructed, and the corresponding phenotypes were analyzed. aceA mutants failed to grow on acetate, and mutants deficient in SMc00767 were also affected in acetate utilization. In contrast, mutants deficient in glcB grew on acetate similar to wild-type strain Rm5000. Complementation experiments showed that aceA and SMc00767 gene constructs were able to restore the growth on acetate in the corresponding single mutants. aceA-glcB, aceA-SMc00767, and glcB-SMc00767 double knockouts were also unable to grow on acetate, but this ability was recovered when the wild-type aceA-glcB or aceA-SMc00767 loci were introduced into the double mutants. These data confirm the functional role of aceA and SMc00767 and show that glcB, in the absence of SMc00767, is required for acetate metabolism. Isocitrate lyase and malate synthase activities were measured in strain Rm5000, the mutant derivatives, and complemented strains. aceA and glcB were able to complement the enzymatic activity lacking in the corresponding single mutants. The enzymatic activities also showed that SMc00767 represses the activity of isocitrate lyase in cells grown on acetate. Gene fusions confirmed the repressor role of SMc00767, which regulates aceA expression at the transcriptional level. Comparison of the transcriptional profiles of the SMc00767 mutant and wild-type strain Rm5000 showed that SMc00767 represses the expression of a moderate number of open reading frames, including aceA; thus, we propose that SMc00767 is a novel repressor involved in acetate metabolism in S. meliloti. Genetic and functional analyses indicated that aceA and SMc00767 constitute a functional two-gene operon, which is conserved in other alpha-proteobacteria. Alfalfa plants infected with the aceA and glcB mutants were not impaired in nodulation or nitrogen fixation, and so the glyoxylate cycle is not required in the Rhizobium-legume symbiosis.
Figures
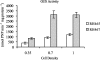
References
-
- Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 62:297-309. - PubMed
-
- Beeckmans, S. April 2001, posting date. Glyoxylate cycle. In Encyclopedia of life sciences. John Wiley & Sons, Ltd., Chichester, United Kingdom. http://www.els.net/.
-
- Beringer, J. E. 1974. R-factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Burris, R. H. 1972. Nitrogen fixation assay—methods and techniques. Methods Enzymol. 24:415-431. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

