Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: a comparison to UV-induced arrest
- PMID: 17526695
- PMCID: PMC1951839
- DOI: 10.1128/JB.00408-07
Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: a comparison to UV-induced arrest
Abstract
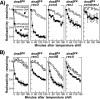
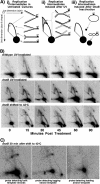
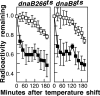
Replication forks face a variety of structurally diverse impediments that can prevent them from completing their task. The mechanism by which cells overcome these hurdles is likely to vary depending on the nature of the obstacle and the strand in which the impediment is encountered. Both UV-induced DNA damage and thermosensitive replication proteins have been used in model systems to inhibit DNA replication and characterize the mechanism by which it recovers. In this study, we examined the molecular events that occur at replication forks following inactivation of a thermosensitive DnaB helicase and found that they are distinct from those that occur following arrest at UV-induced DNA damage. Following UV-induced DNA damage, the integrity of replication forks is maintained and protected from extensive degradation by RecA, RecF, RecO, and RecR until replication can resume. By contrast, inactivation of DnaB results in extensive degradation of the nascent and leading-strand template DNA and a loss of replication fork integrity as monitored by two-dimensional agarose gel analysis. The degradation that occurs following DnaB inactivation partially depends on several genes, including recF, recO, recR, recJ, recG, and xonA. Furthermore, the thermosensitive DnaB allele prevents UV-induced DNA degradation from occurring following arrest even at the permissive temperature, suggesting a role for DnaB prior to loading of the RecFOR proteins. We discuss these observations in relation to potential models for both UV-induced and DnaB(Ts)-mediated replication inhibition.
Figures



References
-
- Bedale, W. A., R. B. Inman, and M. M. Cox. 1993. A reverse DNA strand exchange mediated by recA protein and exonuclease I. The generation of apparent DNA strand breaks by recA protein is explained. J. Biol. Chem. 268:15004-15016. - PubMed
-
- Capaldo, F. N., and S. D. Barbour. 1975. The role of the rec genes in the viability of Escherichia coli K12. Basic Life Sci. 5A:405-418. - PubMed
-
- Carl, P. L. 1970. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet. 109:107-122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

