Chloramphenicol is a substrate for a novel nitroreductase pathway in Haemophilus influenzae
- PMID: 17526758
- PMCID: PMC1932534
- DOI: 10.1128/AAC.00087-07
Chloramphenicol is a substrate for a novel nitroreductase pathway in Haemophilus influenzae
Abstract
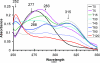
The p-nitroaromatic antibiotic chloramphenicol has been used extensively to treat life-threatening infections due to Haemophilus influenzae and Neisseria meningitidis; its mechanism of action is the inhibition of protein synthesis. We found that during incubation with H. influenzae cells and lysates, chloramphenicol is converted to a 4-aminophenyl allylic alcohol that lacks antibacterial activity. The allylic alcohol moiety undergoes facile re-addition of water to restore the 1,3-diol, as well as further dehydration driven by the aromatic amine to form the iminoquinone. Several Neisseria species and most chloramphenicol-susceptible Haemophilus species, but not Escherichia coli or other gram-negative or gram-positive bacteria we examined, were also found to metabolize chloramphenicol. The products of chloramphenicol metabolism by species other than H. influenzae have not yet been characterized. The strains reducing the antibiotic were chloramphenicol susceptible, indicating that the pathway does not appear to mediate chloramphenicol resistance. The role of this novel nitroreductase pathway in the physiology of H. influenzae and Neisseria species is unknown. Further understanding of the H. influenzae chloramphenicol reduction pathway will contribute to our knowledge of the diversity of prokaryotic nitroreductase mechanisms.
Figures
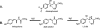

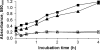


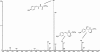
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

