Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation
- PMID: 17531098
- PMCID: PMC1904230
- DOI: 10.1186/1471-2199-8-44
Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation
Abstract
Background: The genetic code imposes a dilemma for cells. The DNA must be long enough to encode for the complexity of an organism, yet thin and flexible enough to fit within the cell. The combination of these properties greatly favors DNA collisions, which can knot and drive recombination of the DNA. Despite the well-accepted propensity of cellular DNA to collide and react with itself, it has not been established what the physiological consequences are.
Results: Here we analyze the effects of recombined and knotted plasmids in E. coli using the Hin site-specific recombination system. We show that Hin-mediated DNA knotting and recombination (i) promote replicon loss by blocking DNA replication; (ii) block gene transcription; and (iii) cause genetic rearrangements at a rate three to four orders of magnitude higher than the rate for an unknotted, unrecombined plasmid.
Conclusion: These results show that DNA reactivity leading to recombined and knotted DNA is potentially toxic and may help drive genetic evolution.
Figures
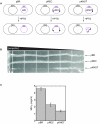

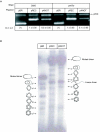
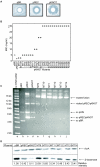
 ), linear dimer (
), linear dimer ( ) catenanes (e.g.,
) catenanes (e.g.,  ) are indicated.
) are indicated.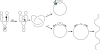
Similar articles
-
Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro.Mol Microbiol. 2004 Feb;51(4):1143-54. doi: 10.1046/j.1365-2958.2003.03890.x. Mol Microbiol. 2004. PMID: 14763986
-
Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli.Biotechnol Bioeng. 2008 Dec 1;101(5):985-95. doi: 10.1002/bit.21976. Biotechnol Bioeng. 2008. PMID: 18553504
-
An Escherichia coli system for assay of F1p site-specific recombination on substrate plasmids.Gene. 1996 Nov 21;180(1-2):225-7. doi: 10.1016/s0378-1119(96)00449-0. Gene. 1996. PMID: 8973372
-
Molecular mechanisms in genetic recombination.Annu Rev Genet. 1973;7:87-111. doi: 10.1146/annurev.ge.07.120173.000511. Annu Rev Genet. 1973. PMID: 4593311 Review. No abstract available.
-
The function and organization of plasmids.Methods Mol Biol. 2003;235:1-17. doi: 10.1385/1-59259-409-3:1. Methods Mol Biol. 2003. PMID: 12904641 Review. No abstract available.
Cited by
-
Molecular Knots.Angew Chem Int Ed Engl. 2017 Sep 4;56(37):11166-11194. doi: 10.1002/anie.201702531. Epub 2017 Aug 16. Angew Chem Int Ed Engl. 2017. PMID: 28477423 Free PMC article. Review.
-
Bullied no more: when and how DNA shoves proteins around.Q Rev Biophys. 2012 Aug;45(3):257-299. doi: 10.1017/S0033583512000054. Epub 2012 Jul 31. Q Rev Biophys. 2012. PMID: 22850561 Free PMC article. Review.
-
Local selection rules that can determine specific pathways of DNA unknotting by type II DNA topoisomerases.Nucleic Acids Res. 2007;35(15):5223-31. doi: 10.1093/nar/gkm532. Epub 2007 Aug 1. Nucleic Acids Res. 2007. PMID: 17670794 Free PMC article.
-
DNA superhelicity.Nucleic Acids Res. 2024 Jan 11;52(1):22-48. doi: 10.1093/nar/gkad1092. Nucleic Acids Res. 2024. PMID: 37994702 Free PMC article.
-
Periodic forces trigger knot untying during translocation of knotted proteins.Sci Rep. 2016 Mar 21;6:21702. doi: 10.1038/srep21702. Sci Rep. 2016. PMID: 26996878 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

