Generation of inhibitor-sensitive protein tyrosine phosphatases via active-site mutations
- PMID: 17532515
- PMCID: PMC1950444
- DOI: 10.1016/j.ymeth.2007.02.005
Generation of inhibitor-sensitive protein tyrosine phosphatases via active-site mutations
Abstract
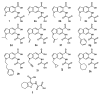
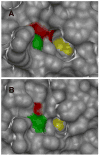
Protein tyrosine phosphatases (PTPs) catalyze the dephosphorylation of phosphotyrosine, a central control element in mammalian signal transduction. Small-molecule inhibitors that are specific for each cellular PTP would be valuable tools in dissecting phosphorylation networks and for validating PTPs as therapeutic targets. However, the common architecture of PTP active sites impedes the discovery of selective PTP inhibitors. Our laboratory has recently used enzyme/inhibitor-interface engineering to generate selective PTP inhibitors. The crux of the strategy resides in the design of "inhibitor-sensitized" PTPs through protein engineering of a novel binding pocket in the target PTP. "Allele-specific" inhibitors that selectively target the sensitized PTP can be synthesized by modifying broad-specificity inhibitors with bulky chemical groups that are incompatible with wild-type PTP active sites; alternatively, specific inhibitors that serendipitously recognize the sensitized PTP's non-natural pocket may be discovered from panels of "non-rationally" designed compounds. In this review, we describe the current state of the PTP-sensitization strategy, with emphases on the methodology of identifying PTP-sensitizing mutations and synthesizing the compounds that have been found to target PTPs in an allele-specific manner. Moreover, we discuss the scope of PTP sensitization in regard to the potential application of the approach across the family of classical PTPs.
Figures
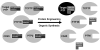

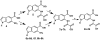
References
-
- Tonks NK. Nat Rev Mol Cell Biol. 2006;7:833–846. - PubMed
-
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Cell. 2004;117:699–711. - PubMed
-
- Tonks NK, Neel BG. Curr Opin Cell Biol. 2001;13:182–195. - PubMed
-
- Van Vactor D, O'Reilly AM, Neel BG. Curr Opin Genet Dev. 1998;8:112–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

