microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos
- PMID: 17532797
- PMCID: PMC2375802
- DOI: 10.1111/j.1469-7580.2007.00746.x
microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos
Abstract
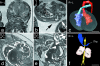
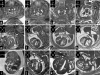
Rapid and precise phenotyping analysis of large numbers of wild-type and mutant mouse embryos is essential for characterizing the genetic and epigenetic factors regulating embryogenesis. We present a novel methodology that permits precise high-throughput screening of the phenotype of embryos with both targeted and randomly generated mutations. To demonstrate the potential of this methodology we show embryo phenotyping results produced in a large-scale ENU-mutagenesis study. In essence this represents an analysis pipeline, which starts with simultaneous micro-magentic resonance imaging (microMRI) screening (voxel size: 25.4 x 25.4 x 24.4 microm) of 32 embryos in one run. Embryos with an indistinct phenotype are then cut into parts and suspect organs and structures are analysed with HREM (high-resolution episcopic microscopy). HREM is an imaging technique that employs 'positive' eosin staining and episcopic imaging for generating three-dimensional (3D) high-resolution (voxel size: 1.07 x 1.07 x 2 microm) digital data of near histological contrast and quality. The results show that our method guarantees the rapid availability of comprehensive phenotype information for high numbers of embryos in, if necessary, histological quality and detail. The combination of high-throughput microMRI with HREM provides an alternative screening pipeline with advantages over existing 3D phenotype screening methods as well as traditional histology. Thus, the microMRI-HREM phenotype analysis pipeline recommends itself as a routine tool for analysing the phenotype of transgenic and mutant embryos.
Figures

References
-
- Bamforth SD, Braganca J, Farthing CR, et al. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. - PubMed
-
- Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium (III) chelates as MRI contrast agents: structure, dynamics, and applications. Chem Rev. 1999;99:2293–2352. - PubMed
-
- Ewald AJ, McBride H, Reddington M, Fraser SE, Kerschmann R. Surface imaging microscopy, an automated method for visualizing whole embryo samples in three dimensions at high resolution. Dev Dyn. 2002;225:369–375. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

