Regulation of plant developmental processes by a novel splicing factor
- PMID: 17534421
- PMCID: PMC1868597
- DOI: 10.1371/journal.pone.0000471
Regulation of plant developmental processes by a novel splicing factor
Abstract
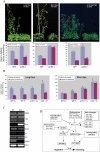
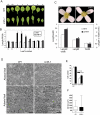
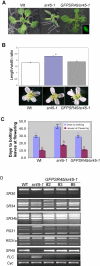
Serine/arginine-rich (SR) proteins play important roles in constitutive and alternative splicing and other aspects of mRNA metabolism. We have previously isolated a unique plant SR protein (SR45) with atypical domain organization. However, the biological and molecular functions of this novel SR protein are not known. Here, we report biological and molecular functions of this protein. Using an in vitro splicing complementation assay, we showed that SR45 functions as an essential splicing factor. Furthermore, the alternative splicing pattern of transcripts of several other SR genes was altered in a mutant, sr45-1, suggesting that the observed phenotypic abnormalities in sr45-1 are likely due to altered levels of SR protein isoforms, which in turn modulate splicing of other pre-mRNAs. sr45-1 exhibited developmental abnormalities, including delayed flowering, narrow leaves and altered number of petals and stamens. The late flowering phenotype was observed under both long days and short days and was rescued by vernalization. FLC, a key flowering repressor, is up-regulated in sr45-1 demonstrating that SR45 influences the autonomous flowering pathway. Changes in the alternative splicing of SR genes and the phenotypic defects in the mutant were rescued by SR45 cDNA, further confirming that the observed defects in the mutant are due to the lack of SR45. These results indicate that SR45 is a novel plant-specific splicing factor that plays a crucial role in regulating developmental processes.
Conflict of interest statement
Figures





References
-
- Reddy AS. Alternative Splicing of Pre-Messenger RNAs in Plants in the Genomic Era. Annu Rev Plant Biol. 2007;58:267–294. - PubMed
-
- Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613–615. - PubMed
-
- Sanford JR, Longman D, Caceres JF. Multiple roles of the SR protein family in splicing regulation. In: Jeanteur P, editor. Regulation of alternative splicing. New York: Springer; 2003. pp. 33–58. - PubMed
-
- Reddy ASN. Nuclear pre-mRNA splicing in plants. CRC Crit Rev Plant Sci. 2001;20:523–572.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

