Global surveillance of emerging Influenza virus genotypes by mass spectrometry
- PMID: 17534439
- PMCID: PMC1876795
- DOI: 10.1371/journal.pone.0000489
Global surveillance of emerging Influenza virus genotypes by mass spectrometry
Abstract
Background: Effective influenza surveillance requires new methods capable of rapid and inexpensive genomic analysis of evolving viral species for pandemic preparedness, to understand the evolution of circulating viral species, and for vaccine strain selection. We have developed one such approach based on previously described broad-range reverse transcription PCR/electrospray ionization mass spectrometry (RT-PCR/ESI-MS) technology.
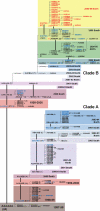
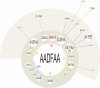
Methods and principal findings: Analysis of base compositions of RT-PCR amplicons from influenza core gene segments (PB1, PB2, PA, M, NS, NP) are used to provide sub-species identification and infer influenza virus H and N subtypes. Using this approach, we detected and correctly identified 92 mammalian and avian influenza isolates, representing 30 different H and N types, including 29 avian H5N1 isolates. Further, direct analysis of 656 human clinical respiratory specimens collected over a seven-year period (1999-2006) showed correct identification of the viral species and subtypes with >97% sensitivity and specificity. Base composition derived clusters inferred from this analysis showed 100% concordance to previously established clades. Ongoing surveillance of samples from the recent influenza virus seasons (2005-2006) showed evidence for emergence and establishment of new genotypes of circulating H3N2 strains worldwide. Mixed viral quasispecies were found in approximately 1% of these recent samples providing a view into viral evolution.
Conclusion/significance: Thus, rapid RT-PCR/ESI-MS analysis can be used to simultaneously identify all species of influenza viruses with clade-level resolution, identify mixed viral populations and monitor global spread and emergence of novel viral genotypes. This high-throughput method promises to become an integral component of influenza surveillance.
Conflict of interest statement
Figures


References
-
- Simonsen L, Fukuda K, Schonberger LB, Cox NJ. The impact of influenza epidemics on hospitalizations. J Infect Dis. 2000;181:831–837. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. - PubMed
-
- Kilbourne ED. Influenza pandemics: can we prepare for the unpredictable? Viral Immunol. 2004;17:350–357. - PubMed
-
- Xu X, Smith CB, Mungall BA, Lindstrom SE, Hall HE, et al. Intercontinental circulation of human influenza A(H1N2) reassortant viruses during the 2001–2002 influenza season. J Infect Dis. 2002;186:1490–1493. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

