Protein structure determination from NMR chemical shifts
- PMID: 17535901
- PMCID: PMC1887584
- DOI: 10.1073/pnas.0610313104
Protein structure determination from NMR chemical shifts
Abstract
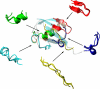
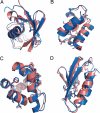
NMR spectroscopy plays a major role in the determination of the structures and dynamics of proteins and other biological macromolecules. Chemical shifts are the most readily and accurately measurable NMR parameters, and they reflect with great specificity the conformations of native and nonnative states of proteins. We show, using 11 examples of proteins representative of the major structural classes and containing up to 123 residues, that it is possible to use chemical shifts as structural restraints in combination with a conventional molecular mechanics force field to determine the conformations of proteins at a resolution of 2 angstroms or better. This strategy should be widely applicable and, subject to further development, will enable quantitative structural analysis to be carried out to address a range of complex biological problems not accessible to current structural techniques.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986.
-
- Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. - PubMed
-
- Wishart DS, Case DA. Methods Enzymol. 2001;338:3–34. - PubMed
-
- Sanders JKM, Hunter BK. Modern NMR Spectroscopy. Oxford, UK: Oxford Univ Press; 1993.
-
- Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647–1651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

