Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR
- PMID: 17535905
- PMCID: PMC1887587
- DOI: 10.1073/pnas.0703820104
Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR
Abstract
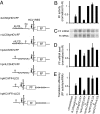
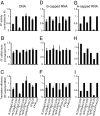
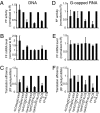
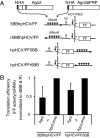
In animals, microRNAs (miRNAs) bind to the 3' UTRs of their target mRNAs and interfere with translation, although the exact mechanism of inhibition of protein synthesis remains unclear. Functional miRNA-binding sites in the coding regions or 5' UTRs of endogenous mRNAs have not been identified. We studied the effect of introducing miRNA target sites into the 5' UTR of luciferase reporter mRNAs containing internal ribosome entry sites (IRESs), so that potential steric hindrance by a microribonucleoprotein complex would not interfere with the initiation of translation. In human HeLa cells, which express endogenous let-7a miRNA, the translational efficiency of these IRES-containing reporters with 5' let-7 complementary sites from the Caenorhabditis elegans lin-41 3' UTR was repressed. Similarly, the IRES-containing reporters were translationally repressed when human Ago2 was tethered to either the 5' or 3' UTR. Interestingly, the method of DNA transfection affected our ability to observe miRNA-mediated repression. Our results suggest that association with any position on a target mRNA is mechanistically sufficient for a microribonucleoprotein to exert repression of translation at some step downstream of initiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Cell. 2005;122:553–563. - PubMed
-
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Science. 2006;312:75–79. - PubMed
-
- Nottrott S, Simard MJ, Richter JD. Nat Struct Mol Biol. 2006;13:1108–1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

