An essential role for the SHIP2-dependent negative feedback loop in neuritogenesis of nerve growth factor-stimulated PC12 cells
- PMID: 17535963
- PMCID: PMC2064282
- DOI: 10.1083/jcb.200609017
An essential role for the SHIP2-dependent negative feedback loop in neuritogenesis of nerve growth factor-stimulated PC12 cells
Abstract
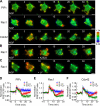
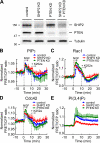
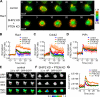
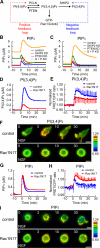
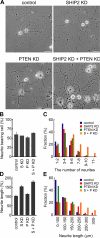
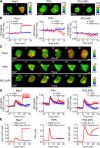
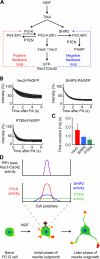
The local accumulation of phosphatidylinositol (3,4,5) trisphosphate (PIP(3)) and resulting activation of Rac1/Cdc42 play a critical role in nerve growth factor (NGF)-induced neurite outgrowth. To further explore the mechanism, we visualized PIP(3), phosphatidylinositol (3,4) bisphosphate, and Rac1/Cdc42 activities by fluorescence resonance energy transfer (FRET) imaging in NGF-stimulated PC12 cells. Based on the obtained FRET images, and with the help of in silico kinetic reaction model, we predicted that PI-5-phosphatase negatively regulates PIP(3) upon NGF stimulation. In agreement with this model, depletion of Src homology 2 domain-containing inositol polyphosphate 5-phosphatase 2 (SHIP2) markedly potentiated NGF-induced Rac1/Cdc42 activation and PIP(3) accumulation and considerably increased the number and the length of neurites in phosphate and tensin homologue-depleted PC12 cells. Further refinement of the computational model predicted Rac1 regulation of PI3-kinase and SHIP2, which was also validated experimentally. We propose that the SHIP2-mediated negative feedback on PIP(3) coordinately works with the PI3-kinase-mediated positive feedback to form an initial protrusive pattern and, later, to punctuate the PIP(3) accumulation to maintain proper neurite outgrowth.
Figures

References
-
- Aoki, K., T. Nakamura, and M. Matsuda. 2004. Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J. Biol. Chem. 279:713–719. - PubMed
-
- Bhalla, U.S. 2004. Models of cell signaling pathways. Curr. Opin. Genet. Dev. 14:375–381. - PubMed
-
- Bourne, H.R., and O. Weiner. 2002. A chemical compass. Nature. 419:21. - PubMed
-
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

