Molecular evidence for a link between the N363S glucocorticoid receptor polymorphism and altered gene expression
- PMID: 17535992
- PMCID: PMC2692663
- DOI: 10.1210/jc.2007-0642
Molecular evidence for a link between the N363S glucocorticoid receptor polymorphism and altered gene expression
Abstract
Context: A single-nucleotide polymorphism (SNP) in the human glucocorticoid receptor (hGR) N363S (rs6195) has been the focus of several clinical studies, and some epidemiological data link this SNP to increased glucocorticoid sensitivity, coronary artery disease, and increased body mass index. However, molecular studies in vitro using reporter gene expression systems have failed, for the most part, to define a link between this polymorphism and altered glucocorticoid receptor function.
Objective: The objective of this study was to address the biological relevancy of N363S SNP in GR function by establishing stable U-2 OS (human osteosarcoma) cell lines expressing wild-type hGR or N363S and examining these receptors under a variety of conditions that probe for GR activity including human gene microarray analysis.
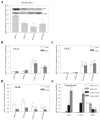
Design: Functional assays with reporter gene systems, Western blotting, and human microarray analysis were used to evaluate the activity of wild-type and N363S GR in both transiently and stably expressing cells. In addition, quantitative RT-PCR was used to confirm the microarray analysis.
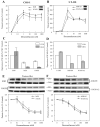
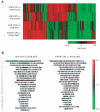
Results: Functional assays with reporter gene systems and homologous down-regulation revealed only minor differences between the wild-type hGR and N363S receptors in both transiently and stably expressing cell lines. However, examination of the two receptors by human gene microarray analysis revealed a unique gene expression profile for N363S.
Conclusions: These studies demonstrate that the N363S SNP regulates a novel set of genes with several of the regulated genes supporting a potential role for this GR polymorphism in human diseases.
Figures

References
-
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. - PubMed
-
- DeRijk RH, Schaaf M, de Kloet ER. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81:103–122. - PubMed
-
- Rosmond R, Dallman MF, Bjorntorp P. Stress-related cortisol secretion in men: relationships with abdominal obesity and endocrine, metabolic, and hemodynamic abnormalities. J Clin Endocrinol Metab. 1998;83:1853–1859. - PubMed
-
- Huizenga NA, Koper JW, de Lange P, Pols HA, Stolk RP, Grobbee DE, de Jong FH, Lamberts SW. Interperson variability but intraperson stability of baseline plasma cortisol concentrations, and its relation to feedback sensitivity of the hypothalamo-pituitary-adrenal axis to a low dose of dexamethasone in elderly individuals. J Clin Endocrinol Metab. 1998;83:47–54. - PubMed
-
- Karl M, Lamberts SW, Detera-Wadleigh SD, Encio IJ, Stratakis CA, Hurley DM, Accili D, Chrousos GP. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J Clin Endocrinol Metab. 1993;76:683–689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

