Requirements and ontology for a G protein-coupled receptor oligomerization knowledge base
- PMID: 17537266
- PMCID: PMC1904246
- DOI: 10.1186/1471-2105-8-177
Requirements and ontology for a G protein-coupled receptor oligomerization knowledge base
Abstract
Background: G Protein-Coupled Receptors (GPCRs) are a large and diverse family of membrane proteins whose members participate in the regulation of most cellular and physiological processes and therefore represent key pharmacological targets. Although several bioinformatics resources support research on GPCRs, most of these have been designed based on the traditional assumption that monomeric GPCRs constitute the functional receptor unit. The increase in the frequency and number of reports about GPCR dimerization/oligomerization and the implication of oligomerization in receptor function makes necessary the ability to store and access information about GPCR dimers/oligomers electronically.
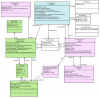
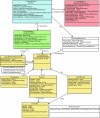
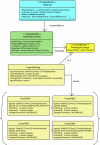
Results: We present here the requirements and ontology (the information scheme to describe oligomers and associated concepts and their relationships) for an information system that can manage the elements of information needed to describe comprehensively the phenomena of both homo- and hetero-oligomerization of GPCRs. The comprehensive information management scheme that we plan to use for the development of an intuitive and user-friendly GPCR-Oligomerization Knowledge Base (GPCR-OKB) is the result of a community dialog involving experimental and computational colleagues working on GPCRs.
Conclusion: Our long term goal is to disseminate to the scientific community organized, curated, and detailed information about GPCR dimerization/oligomerization and its related structural context. This information will be reported as close to the data as possible so the user can make his own judgment on the conclusions drawn for a particular study. The requirements and ontology described here will facilitate the development of future information systems for GPCR oligomers that contain both computational and experimental information about GPCR oligomerization. This information is freely accessible at http://www.gpcr-okb.org.
Figures
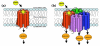

References
-
- Filizola M, Guo W, Javitch JA, Weinstein H. Oligomerization domains of G-protein Coupled Receptors: Insights into the structural basis of GPCR association. In: Devi LA, editor. The G-Protein Coupled Receptor Handbook. Humana Press; 2005. (Contemporary Clinical Neuroscience).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

