Physiological evaluation of the filamentous fungus Trichoderma reesei in production processes by marker gene expression analysis
- PMID: 17537269
- PMCID: PMC1899492
- DOI: 10.1186/1472-6750-7-28
Physiological evaluation of the filamentous fungus Trichoderma reesei in production processes by marker gene expression analysis
Abstract
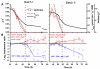
Background: Biologically relevant molecular markers can be used in evaluation of the physiological state of an organism in biotechnical processes. We monitored at high frequency the expression of 34 marker genes in batch, fed-batch and continuous cultures of the filamentous fungus Trichoderma reesei by the transcriptional analysis method TRAC (TRanscript analysis with the aid of Affinity Capture). Expression of specific genes was normalised either with respect to biomass or to overall polyA RNA concentration. Expressional variation of the genes involved in various process relevant cellular functions, such as protein production, growth and stress responses, was related to process parameters such as specific growth and production rates and substrate and dissolved oxygen concentrations.
Results: Gene expression of secreted cellulases and recombinant Melanocarpus albomyces laccase predicted the trends in the corresponding extracellular enzyme production rates and was highest in a narrow "physiological window" in the specific growth rate (micro) range of 0.03-0.05 h-1. Expression of ribosomal protein mRNAs was consistent with the changes in mu. Nine starvation-related genes were found as potential markers for detection of insufficient substrate feed for maintaining optimal protein production. For two genes induced in anaerobic conditions, increasing transcript levels were measured as dissolved oxygen decreased.
Conclusion: The data obtained by TRAC supported the usefulness of focused and intensive transcriptional analysis in monitoring of biotechnical processes providing thus tools for process optimisation purposes.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

