CodonO: codon usage bias analysis within and across genomes
- PMID: 17537810
- PMCID: PMC1933134
- DOI: 10.1093/nar/gkm392
CodonO: codon usage bias analysis within and across genomes
Abstract
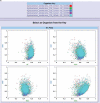
Synonymous codon usage biases are associated with various biological factors, such as gene expression level, gene length, gene translation initiation signal, protein amino acid composition, protein structure, tRNA abundance, mutation frequency and patterns, and GC compositions. Quantification of codon usage bias helps understand evolution of living organisms. A codon usage bias pipeline is demanding for codon usage bias analyses within and across genomes. Here we present a CodonO webserver service as a user-friendly tool for codon usage bias analyses across and within genomes in real time. The webserver is available at http//www.sysbiology.org/CodonO.
Contact: wanhenry@yahoo.com.
Figures

Similar articles
-
Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes.BMC Evol Biol. 2004 Jun 28;4:19. doi: 10.1186/1471-2148-4-19. BMC Evol Biol. 2004. PMID: 15222899 Free PMC article.
-
The mystery of two straight lines in bacterial genome statistics.Bull Math Biol. 2007 Oct;69(7):2429-42. doi: 10.1007/s11538-007-9229-6. Epub 2007 Jun 19. Bull Math Biol. 2007. PMID: 17577600
-
Constraint on di-nucleotides by codon usage bias in bacterial genomes.Gene. 2014 Feb 15;536(1):18-28. doi: 10.1016/j.gene.2013.11.098. Epub 2013 Dec 11. Gene. 2014. PMID: 24333347
-
Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes.Biol Rev Camb Philos Soc. 2013 Feb;88(1):49-61. doi: 10.1111/j.1469-185X.2012.00242.x. Epub 2012 Aug 14. Biol Rev Camb Philos Soc. 2013. PMID: 22889422 Review.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
Cited by
-
Comparative investigation of the various determinants that influence the codon and amino acid usage patterns in the genus Bifidobacterium.World J Microbiol Biotechnol. 2015 Jun;31(6):959-81. doi: 10.1007/s11274-015-1850-1. Epub 2015 Apr 5. World J Microbiol Biotechnol. 2015. PMID: 25842224
-
Why genes evolve faster on secondary chromosomes in bacteria.PLoS Comput Biol. 2010 Apr 1;6(4):e1000732. doi: 10.1371/journal.pcbi.1000732. PLoS Comput Biol. 2010. PMID: 20369015 Free PMC article.
-
Synonymous sites for accessibility around microRNA binding sites in bacterial spot and speck disease resistance genes of tomato.Funct Integr Genomics. 2023 Jul 20;23(3):247. doi: 10.1007/s10142-023-01178-x. Funct Integr Genomics. 2023. PMID: 37468805
-
Gibberellin Oxidase Gene Family in L. chinense: Genome-Wide Identification and Gene Expression Analysis.Int J Mol Sci. 2021 Jul 2;22(13):7167. doi: 10.3390/ijms22137167. Int J Mol Sci. 2021. PMID: 34281216 Free PMC article.
-
Comparative and phylogenetic analysis of chloroplast genomes from four species in Quercus section Cyclobalanopsis.BMC Genom Data. 2024 Jun 10;25(1):57. doi: 10.1186/s12863-024-01232-y. BMC Genom Data. 2024. PMID: 38858616 Free PMC article.
References
-
- Bains W. Codon distribution in vertebrate genes may be used to predict gene length. J. Mol. Biol. 1987;197:379–388. - PubMed
-
- D’Onofrio G, Ghosh TC, Bernardi G. The base composition of the genes is correlated with the secondary structures of the encoded proteins. Gene. 2002;300:179–187. - PubMed
-
- Bernardi G, Bernardi G. Compositional constraints and genome evolution. J. Mol. Evol. 1986;24:1–11. - PubMed
-
- Gu W, Zhou T, Ma J, Sun X, Lu Z. The relationship between synonymous codon usage and protein structure in Escherichia coli and Homo sapiens. Biosystems. 2004;73:89–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

