CodonO: codon usage bias analysis within and across genomes
- PMID: 17537810
- PMCID: PMC1933134
- DOI: 10.1093/nar/gkm392
CodonO: codon usage bias analysis within and across genomes
Abstract
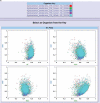
Synonymous codon usage biases are associated with various biological factors, such as gene expression level, gene length, gene translation initiation signal, protein amino acid composition, protein structure, tRNA abundance, mutation frequency and patterns, and GC compositions. Quantification of codon usage bias helps understand evolution of living organisms. A codon usage bias pipeline is demanding for codon usage bias analyses within and across genomes. Here we present a CodonO webserver service as a user-friendly tool for codon usage bias analyses across and within genomes in real time. The webserver is available at http//www.sysbiology.org/CodonO.
Contact: wanhenry@yahoo.com.
Figures

References
-
- Bains W. Codon distribution in vertebrate genes may be used to predict gene length. J. Mol. Biol. 1987;197:379–388. - PubMed
-
- D’Onofrio G, Ghosh TC, Bernardi G. The base composition of the genes is correlated with the secondary structures of the encoded proteins. Gene. 2002;300:179–187. - PubMed
-
- Bernardi G, Bernardi G. Compositional constraints and genome evolution. J. Mol. Evol. 1986;24:1–11. - PubMed
-
- Gu W, Zhou T, Ma J, Sun X, Lu Z. The relationship between synonymous codon usage and protein structure in Escherichia coli and Homo sapiens. Biosystems. 2004;73:89–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

