Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus
- PMID: 17537845
- PMCID: PMC1951341
- DOI: 10.1128/JVI.00690-07
Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus
Abstract
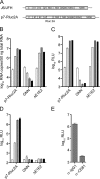
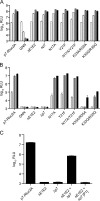
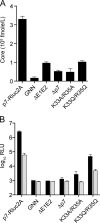
Hepatitis C virus (HCV) infection is a global health concern affecting an estimated 3% of the world's population. Recently, cell culture systems have been established, allowing recapitulation of the complete virus life cycle for the first time. Since the HCV proteins p7 and NS2 are not predicted to be major components of the virion, nor are they required for RNA replication, we investigated whether they might have other roles in the viral life cycle. Here we utilize the recently described infectious J6/JFH chimera to establish that the p7 and NS2 proteins are essential for HCV infectivity. Furthermore, unprocessed forms of p7 and NS2 were not required for this activity. Mutation of two conserved basic residues, previously shown to be important for the ion channel activity of p7 in vitro, drastically impaired infectious virus production. The protease domain of NS2 was required for infectivity, whereas its catalytic active site was dispensable. We conclude that p7 and NS2 function at an early stage of virion morphogenesis, prior to the assembly of infectious virus.
Figures


References
-
- Carrere-Kremer, S., C. Montpellier, L. Lorenzo, B. Brulin, L. Cocquerel, S. Belouzard, F. Penin, and J. Dubuisson. 2004. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 279:41384-41392. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

