Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome
- PMID: 17537853
- PMCID: PMC1951379
- DOI: 10.1128/JVI.00527-07
Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome
Abstract
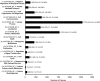
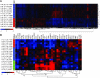
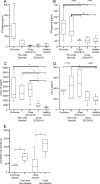
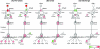
It is not understood how immune inflammation influences the pathogenesis of severe acute respiratory syndrome (SARS). One area of strong controversy is the role of interferon (IFN) responses in the natural history of SARS. The fact that the majority of SARS patients recover after relatively moderate illness suggests that the prevailing notion of deficient type I IFN-mediated immunity, with hypercytokinemia driving a poor clinical course, is oversimplified. We used proteomic and genomic technology to systematically analyze host innate and adaptive immune responses of 40 clinically well-described patients with SARS during discrete phases of illness from the onset of symptoms to discharge or a fatal outcome. A novel signature of high IFN-alpha, IFN-gamma, and IFN-stimulated chemokine levels, plus robust antiviral IFN-stimulated gene (ISG) expression, accompanied early SARS sequelae. As acute illness progressed, SARS patients entered a crisis phase linked to oxygen saturation profiles. The majority of SARS patients resolved IFN responses at crisis and expressed adaptive immune genes. In contrast, patients with poor outcomes showed deviated ISG and immunoglobulin gene expression levels, persistent chemokine levels, and deficient anti-SARS spike antibody production. We contend that unregulated IFN responses during acute-phase SARS may culminate in a malfunction of the switch from innate immunity to adaptive immunity. The potential for the use of the gene signatures we describe in this study to better assess the immunopathology and clinical management of severe viral infections, such as SARS and avian influenza (H5N1), is therefore worth careful examination.
Figures


References
-
- Aubert, M., and K. R. Jerome. 2003. Apoptosis prevention as a mechanism of immune evasion. Int. Rev. Immunol. 22:361-371. - PubMed
-
- Brierley, M. M., and E. N. Fish. 2002. IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 22:835-845. - PubMed
-
- Brierley, M. M., and E. N. Fish. 2005. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 25:733-744. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

