On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization
- PMID: 17540026
- PMCID: PMC1894784
- DOI: 10.1186/1745-6150-2-14
On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization
Abstract
Background: The origin of the translation system is, arguably, the central and the hardest problem in the study of the origin of life, and one of the hardest in all evolutionary biology. The problem has a clear catch-22 aspect: high translation fidelity hardly can be achieved without a complex, highly evolved set of RNAs and proteins but an elaborate protein machinery could not evolve without an accurate translation system. The origin of the genetic code and whether it evolved on the basis of a stereochemical correspondence between amino acids and their cognate codons (or anticodons), through selectional optimization of the code vocabulary, as a "frozen accident" or via a combination of all these routes is another wide open problem despite extensive theoretical and experimental studies. Here we combine the results of comparative genomics of translation system components, data on interaction of amino acids with their cognate codons and anticodons, and data on catalytic activities of ribozymes to develop conceptual models for the origins of the translation system and the genetic code.
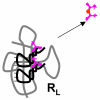
Results: Our main guide in constructing the models is the Darwinian Continuity Principle whereby a scenario for the evolution of a complex system must consist of plausible elementary steps, each conferring a distinct advantage on the evolving ensemble of genetic elements. Evolution of the translation system is envisaged to occur in a compartmentalized ensemble of replicating, co-selected RNA segments, i.e., in a RNA World containing ribozymes with versatile activities. Since evolution has no foresight, the translation system could not evolve in the RNA World as the result of selection for protein synthesis and must have been a by-product of evolution drive by selection for another function, i.e., the translation system evolved via the exaptation route. It is proposed that the evolutionary process that eventually led to the emergence of translation started with the selection for ribozymes binding abiogenic amino acids that stimulated ribozyme-catalyzed reactions. The proposed scenario for the evolution of translation consists of the following steps: binding of amino acids to a ribozyme resulting in an enhancement of its catalytic activity; evolution of the amino-acid-stimulated ribozyme into a peptide ligase (predecessor of the large ribosomal subunit) yielding, initially, a unique peptide activating the original ribozyme and, possibly, other ribozymes in the ensemble; evolution of self-charging proto-tRNAs that were selected, initially, for accumulation of amino acids, and subsequently, for delivery of amino acids to the peptide ligase; joining of the peptide ligase with a distinct RNA molecule (predecessor of the small ribosomal subunit) carrying a built-in template for more efficient, complementary binding of charged proto-tRNAs; evolution of the ability of the peptide ligase to assemble peptides using exogenous RNAs as template for complementary binding of charged proteo-tRNAs, yielding peptides with the potential to activate different ribozymes; evolution of the translocation function of the protoribosome leading to the production of increasingly longer peptides (the first proteins), i.e., the origin of translation. The specifics of the recognition of amino acids by proto-tRNAs and the origin of the genetic code depend on whether or not there is a physical affinity between amino acids and their cognate codons or anticodons, a problem that remains unresolved.
Conclusion: We describe a stepwise model for the origin of the translation system in the ancient RNA world such that each step confers a distinct advantage onto an ensemble of co-evolving genetic elements. Under this scenario, the primary cause for the emergence of translation was the ability of amino acids and peptides to stimulate reactions catalyzed by ribozymes. Thus, the translation system might have evolved as the result of selection for ribozymes capable of, initially, efficient amino acid binding, and subsequently, synthesis of increasingly versatile peptides. Several aspects of this scenario are amenable to experimental testing.
Figures



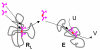

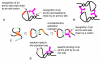
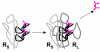

References
-
- Darwin C. On the Origin of Species by Means of Natural Selection or, The Preservation of Races in the Struggle for Life. London , John Murray; 1859.
LinkOut - more resources
Full Text Sources
Other Literature Sources

