Genome-level analysis of genetic regulation of liver gene expression networks
- PMID: 17542012
- PMCID: PMC3518845
- DOI: 10.1002/hep.21682
Genome-level analysis of genetic regulation of liver gene expression networks
Abstract
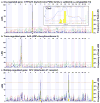
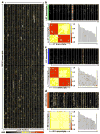
The liver is the primary site for the metabolism of nutrients, drugs, and chemical agents. Although metabolic pathways are complex and tightly regulated, genetic variation among individuals, reflected in variations in gene expression levels, introduces complexity into research on liver disease. This study dissected genetic networks that control liver gene expression through the combination of large-scale quantitative mRNA expression analysis with genetic mapping in a reference population of BXD recombinant inbred mouse strains for which extensive single-nucleotide polymorphism, haplotype, and phenotypic data are publicly available. We profiled gene expression in livers of naive mice of both sexes from C57BL/6J, DBA/2J, B6D2F1, and 37 BXD strains using Agilent oligonucleotide microarrays. These data were used to map quantitative trait loci (QTLs) responsible for variations in the expression of about 19,000 transcripts. We identified polymorphic local and distant QTLs, including several loci that control the expression of large numbers of genes in liver, by comparing the physical transcript position with the location of the controlling QTL.
Conclusion: The data are available through a public web-based resource (www.genenetwork.org) that allows custom data mining, identification of coregulated transcripts and correlated phenotypes, cross-tissue, and cross-species comparisons, as well as testing of a broad array of hypotheses.
Conflict of interest statement
The authors declare they have no competing financial interest.
Figures




References
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
-
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
-
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. - PubMed
-
- Waring JF, Jolly RA, Ciurlionis R, Lum PY, Praestgaard JT, Morfitt DC, et al. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol Appl Pharmacol. 2001;175:28–42. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
