Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss
- PMID: 17542646
- PMCID: PMC1885273
- DOI: 10.1371/journal.ppat.0030079
Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss
Abstract
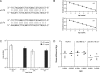
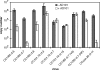
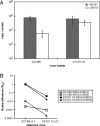
Fitness is a parameter used to quantify how well an organism adapts to its environment; in the present study, fitness is a measure of how well strains of human immunodeficiency virus type 1 (HIV-1) replicate in tissue culture. When HIV-1 develops resistance in vitro or in vivo to antiretroviral drugs such as reverse transcriptase or protease inhibitors, its fitness is often impaired. Here, we have investigated whether the development of resistance in vitro to a small molecule CCR5 inhibitor, AD101, has an associated fitness cost. To do this, we developed a growth-competition assay involving dual infections with molecularly cloned viruses that are essentially isogenic outside the env genes under study. Real-time TaqMan quantitative PCR (QPCR) was used to quantify each competing virus individually via probes specific to different, phenotypically silent target sequences engineered within their vif genes. Head-to-head competition assays of env clones derived from the AD101 escape mutant isolate, the inhibitor-sensitive parental virus, and a passage control virus showed that AD101 resistance was not associated with a fitness loss. This observation is consistent with the retention of the resistant phenotype when the escape mutant was cultured for a total of 20 passages in the absence of the selecting compound. Amino acid substitutions in the V3 region of gp120 that confer complete AD101 resistance cause a fitness loss when introduced into an AD101-sensitive, parental clone; however, in the resistant isolate, changes elsewhere in env that occurred prior to the substitutions within V3 appear to compensate for the adverse effect of the V3 changes on replicative capacity. These in vitro studies may have implications for the development and management of resistance to other CCR5 inhibitors that are being evaluated clinically for the treatment of HIV-1 infection.
Conflict of interest statement
Figures




References
-
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. - PubMed
-
- Quinones-Mateu ME, Arts EJ. Virus fitness: Concept, quantification, and application to HIV population dynamics. Curr Top Microbiol Immunol. 2006;299:83–140. - PubMed
-
- Nijhuis M, Deeks S, Boucher C. Implications of antiretroviral resistance on viral fitness. Curr Opin Infect Dis. 2001;14:23–28. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

