Whole-genome cartography of estrogen receptor alpha binding sites
- PMID: 17542648
- PMCID: PMC1885282
- DOI: 10.1371/journal.pgen.0030087
Whole-genome cartography of estrogen receptor alpha binding sites
Abstract
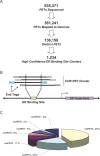
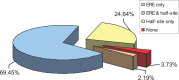
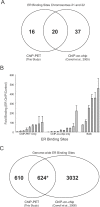
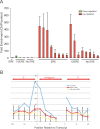
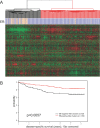
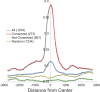
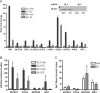
Using a chromatin immunoprecipitation-paired end diTag cloning and sequencing strategy, we mapped estrogen receptor alpha (ERalpha) binding sites in MCF-7 breast cancer cells. We identified 1,234 high confidence binding clusters of which 94% are projected to be bona fide ERalpha binding regions. Only 5% of the mapped estrogen receptor binding sites are located within 5 kb upstream of the transcriptional start sites of adjacent genes, regions containing the proximal promoters, whereas vast majority of the sites are mapped to intronic or distal locations (>5 kb from 5' and 3' ends of adjacent transcript), suggesting transcriptional regulatory mechanisms over significant physical distances. Of all the identified sites, 71% harbored putative full estrogen response elements (EREs), 25% bore ERE half sites, and only 4% had no recognizable ERE sequences. Genes in the vicinity of ERalpha binding sites were enriched for regulation by estradiol in MCF-7 cells, and their expression profiles in patient samples segregate ERalpha-positive from ERalpha-negative breast tumors. The expression dynamics of the genes adjacent to ERalpha binding sites suggest a direct induction of gene expression through binding to ERE-like sequences, whereas transcriptional repression by ERalpha appears to be through indirect mechanisms. Our analysis also indicates a number of candidate transcription factor binding sites adjacent to occupied EREs at frequencies much greater than by chance, including the previously reported FOXA1 sites, and demonstrate the potential involvement of one such putative adjacent factor, Sp1, in the global regulation of ERalpha target genes. Unexpectedly, we found that only 22%-24% of the bona fide human ERalpha binding sites were overlapping conserved regions in whole genome vertebrate alignments, which suggest limited conservation of functional binding sites. Taken together, this genome-scale analysis suggests complex but definable rules governing ERalpha binding and gene regulation.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



References
-
- Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, et al. Effects of estrogen on global gene expression: Identification of novel targets of estrogen action. Cancer Res. 2000;60:5977–5983. - PubMed
-
- Finlin BS, Gau CL, Murphy GA, Shao H, Kimel T, et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J Biol Chem. 2001;276:42259–42267. - PubMed
-
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, et al. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. - PubMed
-
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. - PubMed
-
- Inoue A, Yoshida N, Omoto Y, Oguchi S, Yamori T, et al. Development of cDNA microarray for expression profiling of estrogen-responsive genes. J Mol Endocrinol. 2002;29:175–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

